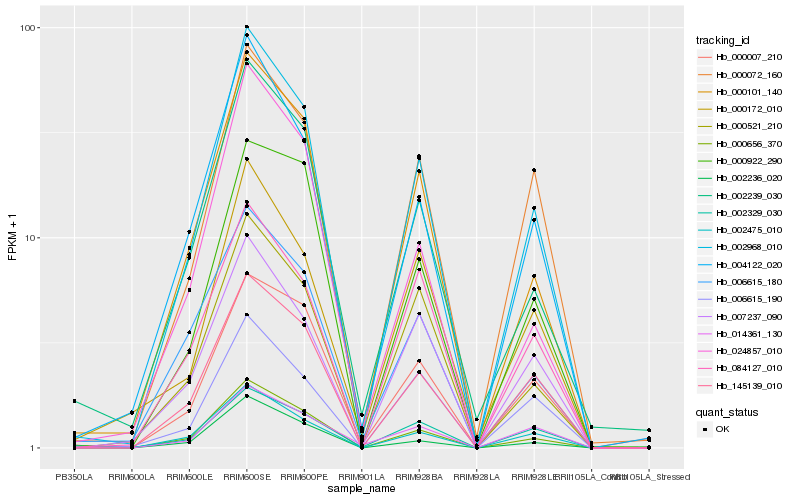
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002968_010 |
0.0 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 2 |
Hb_000656_370 |
0.0568014011 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000101_140 |
0.0744197146 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_145139_010 |
0.07587028 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 isoform X1 [Vitis vinifera] |
| 5 |
Hb_004122_020 |
0.084204992 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 6 |
Hb_000072_160 |
0.0848632039 |
- |
- |
proline oxidase [Manihot esculenta] |
| 7 |
Hb_002475_010 |
0.101505959 |
- |
- |
hypothetical protein JCGZ_22530 [Jatropha curcas] |
| 8 |
Hb_002236_020 |
0.1055925464 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 9 |
Hb_000521_210 |
0.1059773996 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 10 |
Hb_002239_030 |
0.1062021081 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 11 |
Hb_007237_090 |
0.1080160583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002329_030 |
0.1143783598 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000007_210 |
0.1144217743 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 14 |
Hb_006615_180 |
0.1161035402 |
- |
- |
hypothetical protein B456_003G103400 [Gossypium raimondii] |
| 15 |
Hb_006615_190 |
0.1176323328 |
- |
- |
UDP-glycosyltransferase 1 [Linum usitatissimum] |
| 16 |
Hb_000172_010 |
0.1178390463 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670-like isoform X1 [Populus euphratica] |
| 17 |
Hb_084127_010 |
0.1181503835 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_024857_010 |
0.1187317108 |
- |
- |
glutaredoxin 2.1 [Hevea brasiliensis] |
| 19 |
Hb_000922_290 |
0.118943329 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 20 |
Hb_014361_130 |
0.1200535169 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |