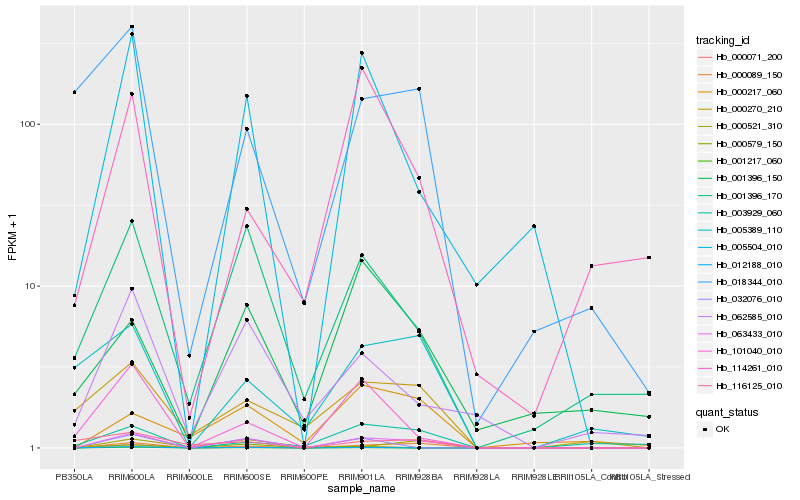
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063433_010 |
0.0 |
- |
- |
Chloroplast channel forming outer membrane protein isoform 2 [Theobroma cacao] |
| 2 |
Hb_000089_150 |
0.1737019653 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 3 |
Hb_005504_010 |
0.2441235978 |
- |
- |
hypothetical protein POPTR_1605s00200g [Populus trichocarpa] |
| 4 |
Hb_001396_170 |
0.2776597004 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_101040_010 |
0.2986894981 |
- |
- |
- |
| 6 |
Hb_032076_010 |
0.3031579252 |
- |
- |
- |
| 7 |
Hb_000217_060 |
0.3069366418 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_062585_010 |
0.3088671503 |
- |
- |
hypothetical protein JCGZ_19532 [Jatropha curcas] |
| 9 |
Hb_012188_010 |
0.3127988064 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 10 |
Hb_000270_210 |
0.3134681947 |
- |
- |
PREDICTED: transcription initiation factor IIB-2-like [Jatropha curcas] |
| 11 |
Hb_001217_060 |
0.3199879658 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 12 |
Hb_001396_150 |
0.3238795046 |
transcription factor |
TF Family: M-type |
hypothetical protein POPTR_0001s13660g [Populus trichocarpa] |
| 13 |
Hb_003929_060 |
0.3253403583 |
- |
- |
PREDICTED: uncharacterized protein ECU03_1610 [Jatropha curcas] |
| 14 |
Hb_000521_310 |
0.3287153108 |
- |
- |
- |
| 15 |
Hb_000071_200 |
0.3335906627 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 16 |
Hb_114261_010 |
0.3354666559 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 17 |
Hb_005389_110 |
0.3374100365 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 18 |
Hb_018344_010 |
0.3457869909 |
- |
- |
PREDICTED: endochitinase 2-like [Populus euphratica] |
| 19 |
Hb_000579_150 |
0.3495121981 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 20 |
Hb_116125_010 |
0.3568703772 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710-like [Citrus sinensis] |