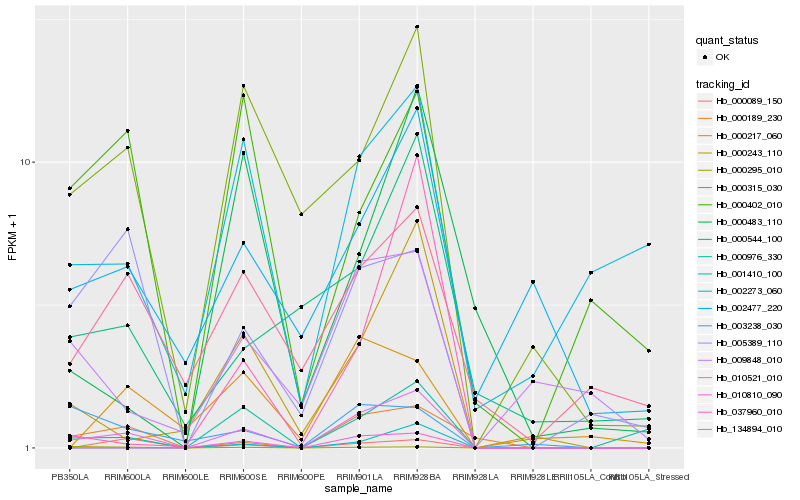
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010521_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 2 |
Hb_000189_230 |
0.262669578 |
- |
- |
Auxin-induced protein [Aegilops tauschii] |
| 3 |
Hb_000976_330 |
0.2689855616 |
- |
- |
hypothetical protein POPTR_0001s05180g [Populus trichocarpa] |
| 4 |
Hb_000243_110 |
0.2696095447 |
- |
- |
PREDICTED: WAT1-related protein At3g53210-like [Jatropha curcas] |
| 5 |
Hb_000315_030 |
0.2902700471 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 6 |
Hb_010810_090 |
0.2949940475 |
- |
- |
- |
| 7 |
Hb_000089_150 |
0.3128772223 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 8 |
Hb_005389_110 |
0.3202397102 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 9 |
Hb_134894_010 |
0.3219511328 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 10 |
Hb_002477_220 |
0.3227488507 |
- |
- |
hypothetical protein POPTR_0017s06650g [Populus trichocarpa] |
| 11 |
Hb_001410_100 |
0.3233767626 |
- |
- |
hypothetical protein EUGRSUZ_B00966 [Eucalyptus grandis] |
| 12 |
Hb_000544_100 |
0.3235346845 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 13 |
Hb_003238_030 |
0.3290957238 |
- |
- |
hypothetical protein EUGRSUZ_I00356 [Eucalyptus grandis] |
| 14 |
Hb_009848_010 |
0.3320170776 |
- |
- |
putative acyl-activating enzyme 16, chloroplastic -like protein [Gossypium arboreum] |
| 15 |
Hb_002273_060 |
0.3323161807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_037960_010 |
0.3355251459 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000217_060 |
0.3393431273 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000483_110 |
0.3422084205 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 2, partial [Theobroma cacao] |
| 19 |
Hb_000295_010 |
0.3433910155 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_000402_010 |
0.3448445592 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 6 [Jatropha curcas] |