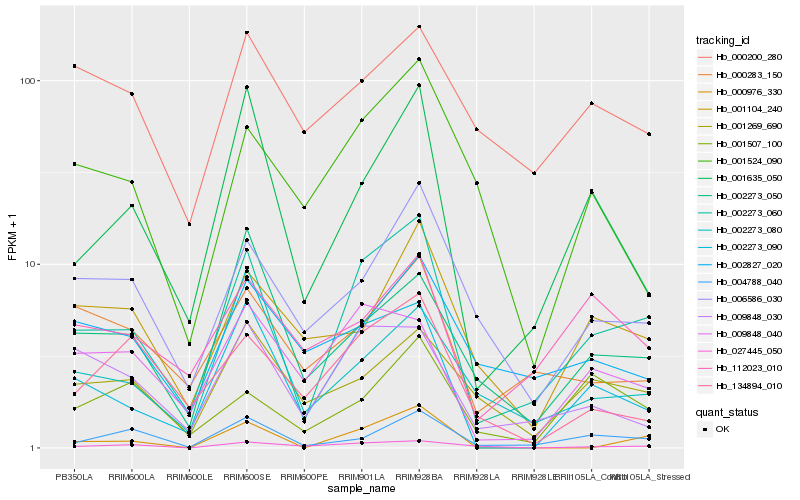
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002273_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002273_090 |
0.1769751285 |
- |
- |
- |
| 3 |
Hb_001635_050 |
0.1979736573 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Nicotiana sylvestris] |
| 4 |
Hb_004788_040 |
0.2029417519 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_006586_030 |
0.2100510277 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 6 |
Hb_009848_030 |
0.2139116639 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 7 |
Hb_009848_040 |
0.2176551038 |
- |
- |
hypothetical protein B456_009G349600 [Gossypium raimondii] |
| 8 |
Hb_000283_150 |
0.2250045352 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 9 |
Hb_001269_690 |
0.2281704377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_112023_010 |
0.2307269811 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 11 |
Hb_001104_240 |
0.2334147974 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000976_330 |
0.233551288 |
- |
- |
hypothetical protein POPTR_0001s05180g [Populus trichocarpa] |
| 13 |
Hb_002273_050 |
0.233954824 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_002827_020 |
0.2344168648 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 15 |
Hb_134894_010 |
0.2365845922 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_027445_050 |
0.2378508114 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |
| 17 |
Hb_002273_080 |
0.239649856 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 18 |
Hb_001507_100 |
0.2422838792 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_001524_090 |
0.2442632886 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 20 |
Hb_000200_280 |
0.2484507378 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |