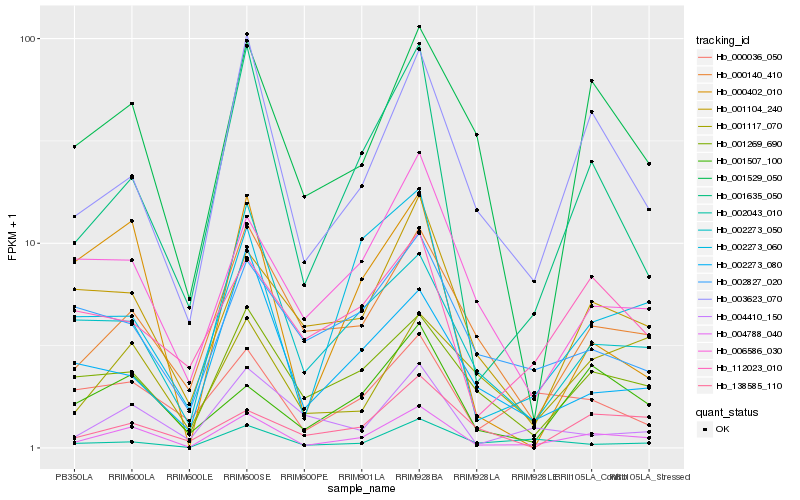
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004788_040 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_003623_070 |
0.1586041811 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_001635_050 |
0.1676833779 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Nicotiana sylvestris] |
| 4 |
Hb_001269_690 |
0.1874366648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000140_410 |
0.1875676495 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001529_050 |
0.1932324238 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 7 |
Hb_001104_240 |
0.1965339761 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_001507_100 |
0.1971339469 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_002273_060 |
0.2029417519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006586_030 |
0.2070928197 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 11 |
Hb_002273_050 |
0.2079421959 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_001117_070 |
0.2130808503 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 13 |
Hb_000036_050 |
0.2183486549 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |
| 14 |
Hb_004410_150 |
0.2186103824 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 15 |
Hb_002043_010 |
0.2186789015 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_138585_110 |
0.2246486457 |
- |
- |
- |
| 17 |
Hb_112023_010 |
0.2249546626 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 18 |
Hb_002827_020 |
0.2282650386 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 19 |
Hb_002273_080 |
0.2285903712 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 20 |
Hb_000402_010 |
0.2324232545 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 6 [Jatropha curcas] |