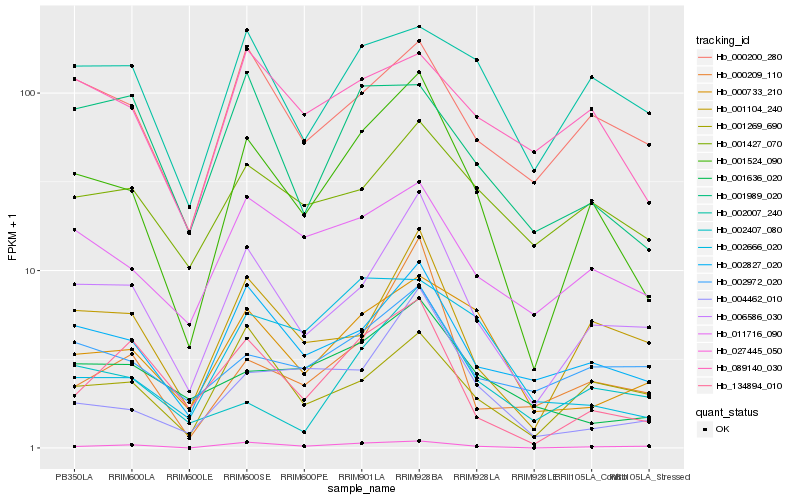
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001524_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 2 |
Hb_006586_030 |
0.1308260247 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 3 |
Hb_002827_020 |
0.1649165456 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 4 |
Hb_001104_240 |
0.1755292055 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000733_210 |
0.1841440598 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 6 |
Hb_027445_050 |
0.1873516873 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |
| 7 |
Hb_134894_010 |
0.1906748432 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_011716_090 |
0.1933806534 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 9 |
Hb_004462_010 |
0.1953303468 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 10 |
Hb_000200_280 |
0.1993807214 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 11 |
Hb_001269_690 |
0.2017174799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002972_020 |
0.2040189893 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 13 |
Hb_001636_020 |
0.2052743473 |
- |
- |
Serine/threonine-protein kinase PBS1 [Theobroma cacao] |
| 14 |
Hb_001427_070 |
0.2085227239 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 15 |
Hb_089140_030 |
0.2121973589 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 16 |
Hb_002007_240 |
0.2126097405 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 17 |
Hb_002407_080 |
0.2135067 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 18 |
Hb_000209_110 |
0.2188567594 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 19 |
Hb_002666_020 |
0.2190891718 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 20 |
Hb_001989_020 |
0.2221279288 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |