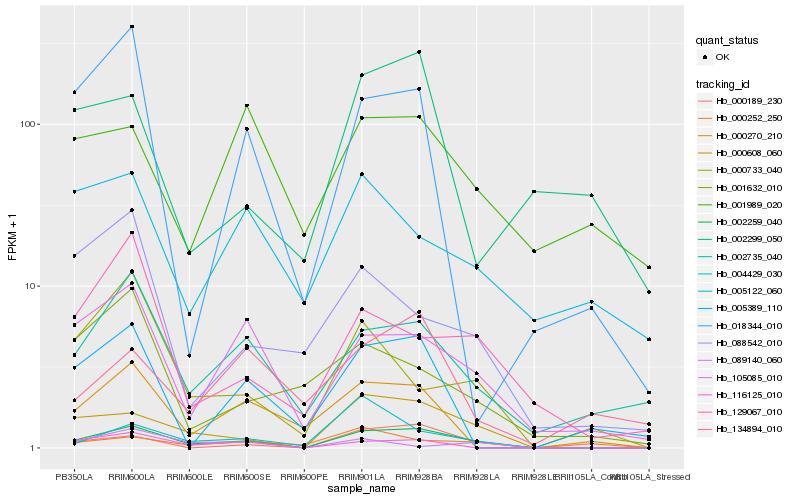
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_040 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000189_230 |
0.2176145732 |
- |
- |
Auxin-induced protein [Aegilops tauschii] |
| 3 |
Hb_002735_040 |
0.2397814704 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 4 |
Hb_105085_010 |
0.2405414721 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 5 |
Hb_116125_010 |
0.245942886 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710-like [Citrus sinensis] |
| 6 |
Hb_000608_060 |
0.2578229744 |
- |
- |
- |
| 7 |
Hb_000270_210 |
0.2697364666 |
- |
- |
PREDICTED: transcription initiation factor IIB-2-like [Jatropha curcas] |
| 8 |
Hb_000733_040 |
0.2738952259 |
- |
- |
hypothetical protein POPTR_0010s20310g [Populus trichocarpa] |
| 9 |
Hb_129067_010 |
0.286543037 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 10 |
Hb_004429_030 |
0.2915414357 |
- |
- |
hypothetical protein POPTR_0010s16560g [Populus trichocarpa] |
| 11 |
Hb_005389_110 |
0.2926552597 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 12 |
Hb_018344_010 |
0.2927500956 |
- |
- |
PREDICTED: endochitinase 2-like [Populus euphratica] |
| 13 |
Hb_000252_250 |
0.2971504364 |
- |
- |
unknown [Lotus japonicus] |
| 14 |
Hb_134894_010 |
0.2986334545 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_089140_060 |
0.308118465 |
- |
- |
Uncharacterized protein TCM_037529 [Theobroma cacao] |
| 16 |
Hb_088542_010 |
0.3086409928 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 17 |
Hb_001632_010 |
0.3101955244 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 18 |
Hb_002299_050 |
0.3109021504 |
- |
- |
PREDICTED: chalcone--flavonone isomerase isoform X1 [Pyrus x bretschneideri] |
| 19 |
Hb_005122_060 |
0.3109177356 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001989_020 |
0.313451255 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |