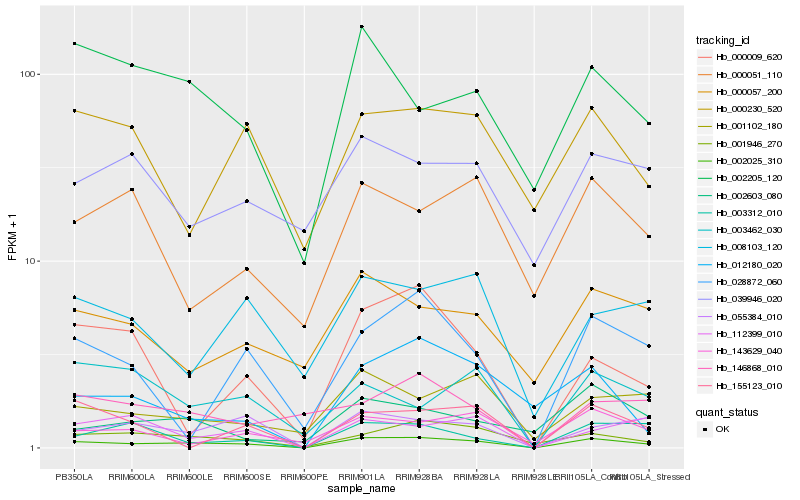
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_310 |
0.0 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_001946_270 |
0.1970457736 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 3 |
Hb_055384_010 |
0.1997155956 |
- |
- |
- |
| 4 |
Hb_001102_180 |
0.2187240252 |
- |
- |
eukaryotic translation initiation factor, putative [Ricinus communis] |
| 5 |
Hb_002205_120 |
0.2213944695 |
- |
- |
PREDICTED: O-acetyl-ADP-ribose deacetylase MACROD2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_028872_060 |
0.2227480968 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 7 |
Hb_003312_010 |
0.2273305665 |
- |
- |
PREDICTED: uncharacterized protein LOC104428797 [Eucalyptus grandis] |
| 8 |
Hb_008103_120 |
0.227387958 |
- |
- |
hypothetical protein CISIN_1g009650mg [Citrus sinensis] |
| 9 |
Hb_143629_040 |
0.2279237644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000057_200 |
0.2283698309 |
- |
- |
PREDICTED: methyltransferase-like protein 22 [Jatropha curcas] |
| 11 |
Hb_000230_520 |
0.2340920839 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 12 |
Hb_112399_010 |
0.2345281226 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_000009_620 |
0.2355670201 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_003462_030 |
0.2371063271 |
- |
- |
hypothetical protein UMN_5210_01, partial [Pinus radiata] |
| 15 |
Hb_002603_080 |
0.238635004 |
- |
- |
hypothetical protein MTR_7g114400 [Medicago truncatula] |
| 16 |
Hb_146868_010 |
0.2388980174 |
- |
- |
- |
| 17 |
Hb_012180_020 |
0.2389684367 |
- |
- |
hypothetical protein POPTR_0001s15100g [Populus trichocarpa] |
| 18 |
Hb_000051_110 |
0.2400473067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_155123_010 |
0.2404379076 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 20 |
Hb_039946_020 |
0.2406834786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |