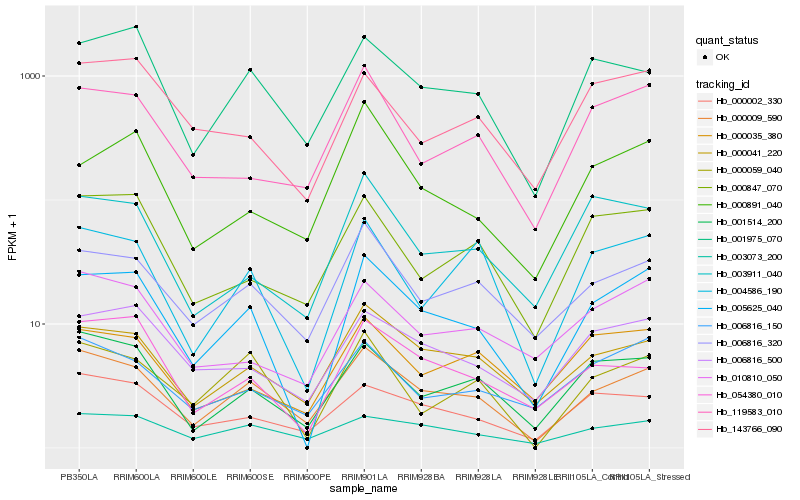
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005625_040 |
0.0 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 2 |
Hb_000009_590 |
0.1308140523 |
- |
- |
PREDICTED: prolyl endopeptidase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_006816_500 |
0.1505856362 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 4 |
Hb_000041_220 |
0.1517654652 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 5 |
Hb_001514_200 |
0.156894208 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 6 |
Hb_004586_190 |
0.1575061607 |
- |
- |
PREDICTED: protein CutA, chloroplastic [Jatropha curcas] |
| 7 |
Hb_006816_320 |
0.1591706696 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Jatropha curcas] |
| 8 |
Hb_001975_070 |
0.1663786679 |
- |
- |
eukaryotic translation elongation factor 1B alpha-subunit [Hevea brasiliensis] |
| 9 |
Hb_000059_040 |
0.1670010434 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 10 |
Hb_119583_010 |
0.1673818538 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 11 |
Hb_003073_200 |
0.1685125991 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 12 |
Hb_000002_330 |
0.1699560926 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 13 |
Hb_003911_040 |
0.169966957 |
- |
- |
PREDICTED: putative H/ACA ribonucleoprotein complex subunit 1-like protein 1 [Jatropha curcas] |
| 14 |
Hb_006816_150 |
0.1702264445 |
- |
- |
PREDICTED: lon protease homolog 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_054380_010 |
0.1705051466 |
- |
- |
- |
| 16 |
Hb_000891_040 |
0.1708381997 |
- |
- |
hypothetical protein JCGZ_11252 [Jatropha curcas] |
| 17 |
Hb_000035_380 |
0.1716194179 |
- |
- |
- |
| 18 |
Hb_143766_090 |
0.171679929 |
- |
- |
60S ribosomal protein L31B [Hevea brasiliensis] |
| 19 |
Hb_010810_050 |
0.1730443014 |
- |
- |
hypothetical protein [Cleome spinosa] |
| 20 |
Hb_000847_070 |
0.1731571231 |
- |
- |
PREDICTED: uncharacterized protein LOC105125699 [Populus euphratica] |