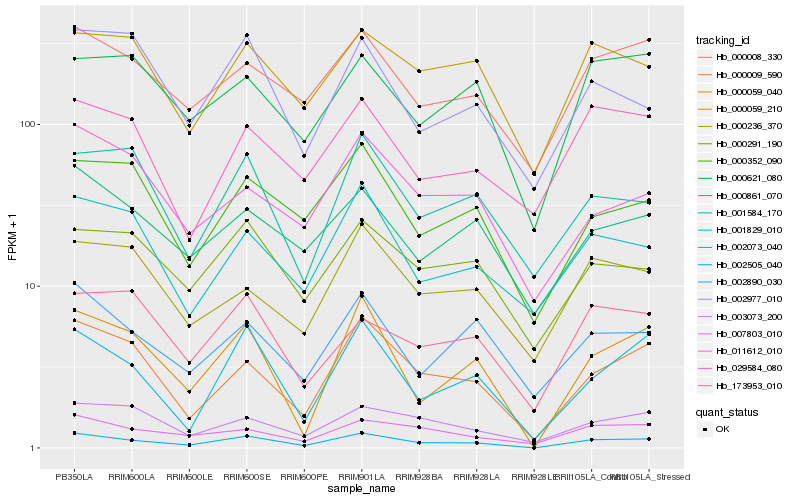
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002505_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000009_590 |
0.1191758345 |
- |
- |
PREDICTED: prolyl endopeptidase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000059_040 |
0.126990089 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 4 |
Hb_002073_040 |
0.1328035824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000008_330 |
0.1480169388 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 6 |
Hb_007803_010 |
0.1502312903 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 7 |
Hb_003073_200 |
0.1511702358 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 8 |
Hb_000352_090 |
0.1537519473 |
- |
- |
PREDICTED: syntaxin-22-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002890_030 |
0.1540277272 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 10 |
Hb_001829_010 |
0.1553277609 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000291_190 |
0.1566651453 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 12 |
Hb_000059_210 |
0.1568963735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001584_170 |
0.1586250672 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 14 |
Hb_000861_070 |
0.1588870659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000236_370 |
0.1620931293 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 16 |
Hb_173953_010 |
0.1623999693 |
- |
- |
hypothetical protein RCOM_1267370 [Ricinus communis] |
| 17 |
Hb_000621_080 |
0.1635009213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011612_010 |
0.1646265202 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 19 |
Hb_002977_010 |
0.1651062273 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 20 |
Hb_029584_080 |
0.1653029067 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |