| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
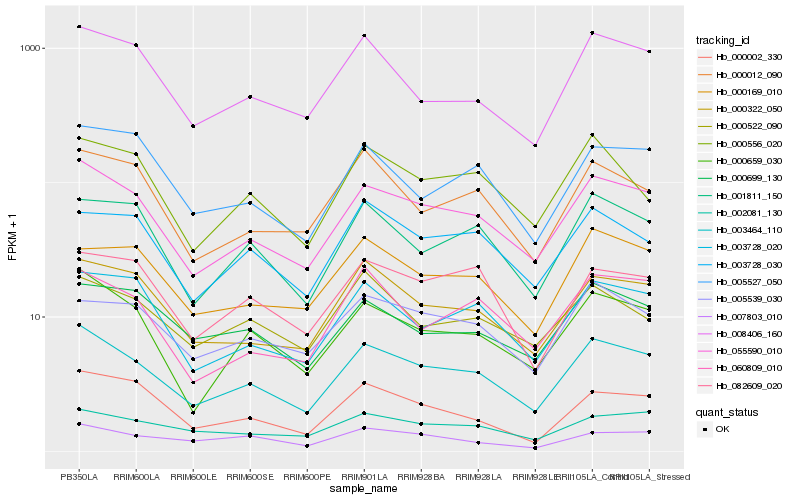
Hb_003464_110 |
0.0 |
- |
- |
3-deoxy-d-manno-octulosonic-acid transferase, putative [Ricinus communis] |
| 2 |
Hb_055590_010 |
0.0381010926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000659_030 |
0.0843147578 |
- |
- |
pollen Ole e 1 allergen and extensin family protein, partial [Manihot esculenta] |
| 4 |
Hb_000322_050 |
0.089767611 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 5 |
Hb_008406_160 |
0.0969849953 |
- |
- |
PREDICTED: ADP,ATP carrier protein 3, mitochondrial [Jatropha curcas] |
| 6 |
Hb_003728_020 |
0.099361698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001811_150 |
0.0994625313 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000012_090 |
0.1049449403 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 9 |
Hb_002081_130 |
0.1053603136 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 10 |
Hb_000699_130 |
0.1059073061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005539_030 |
0.1060896955 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 12 |
Hb_003728_030 |
0.1078859979 |
- |
- |
o-sialoglycoprotein endopeptidase, putative [Ricinus communis] |
| 13 |
Hb_000556_020 |
0.1084184375 |
- |
- |
PREDICTED: terminal uridylyltransferase 7 [Jatropha curcas] |
| 14 |
Hb_000002_330 |
0.1086552398 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 15 |
Hb_005527_050 |
0.1089595279 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |
| 16 |
Hb_007803_010 |
0.1092239221 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 17 |
Hb_060809_010 |
0.1095872632 |
- |
- |
PREDICTED: uncharacterized protein LOC105163892 [Sesamum indicum] |
| 18 |
Hb_000169_010 |
0.1099053993 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_082609_020 |
0.1107898826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000522_090 |
0.1116824241 |
- |
- |
PREDICTED: actin-related protein 9 isoform X1 [Nicotiana tomentosiformis] |