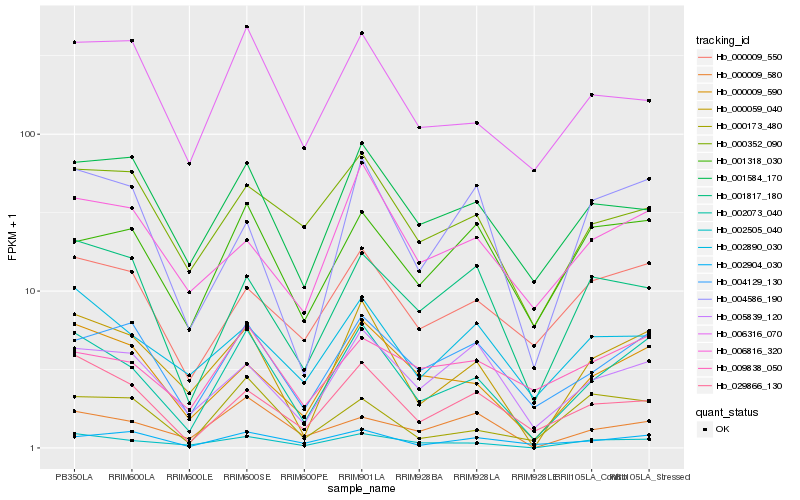
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002073_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000059_040 |
0.1277757794 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 3 |
Hb_002505_040 |
0.1328035824 |
- |
- |
- |
| 4 |
Hb_000009_590 |
0.1453946788 |
- |
- |
PREDICTED: prolyl endopeptidase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004129_130 |
0.1557717114 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_029866_130 |
0.169292802 |
- |
- |
hypothetical protein JCGZ_00035 [Jatropha curcas] |
| 7 |
Hb_000009_550 |
0.1730370113 |
- |
- |
hypothetical protein RCOM_0603640 [Ricinus communis] |
| 8 |
Hb_002904_030 |
0.1737516283 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420 [Jatropha curcas] |
| 9 |
Hb_001318_030 |
0.1778075824 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 10 |
Hb_004586_190 |
0.1782988265 |
- |
- |
PREDICTED: protein CutA, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001584_170 |
0.1794879662 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 12 |
Hb_005839_120 |
0.1799573174 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 13 |
Hb_001817_180 |
0.1819224978 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 14 |
Hb_006316_070 |
0.1823241107 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 15 |
Hb_009838_050 |
0.1826707467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000009_580 |
0.18288323 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 17 |
Hb_002890_030 |
0.1830915938 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 18 |
Hb_000173_480 |
0.1844813137 |
- |
- |
PREDICTED: calcium-binding protein 39 [Jatropha curcas] |
| 19 |
Hb_000352_090 |
0.1855035224 |
- |
- |
PREDICTED: syntaxin-22-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_006816_320 |
0.1855320488 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Jatropha curcas] |