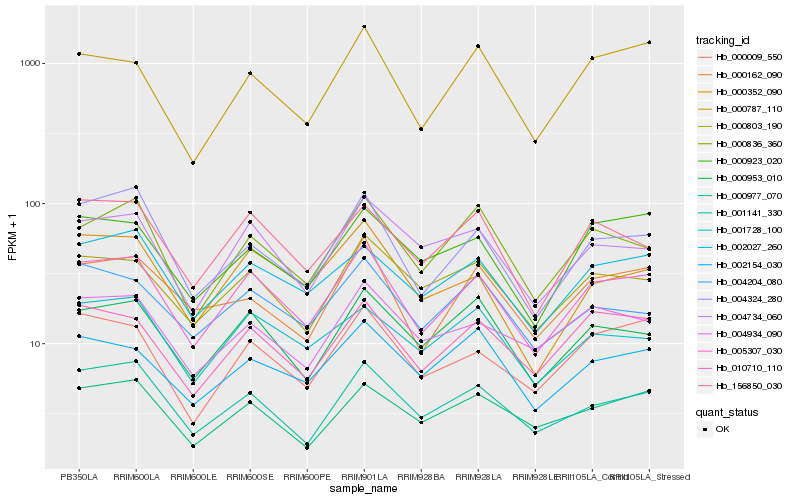
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005307_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s04640g [Populus trichocarpa] |
| 2 |
Hb_010710_110 |
0.0776827399 |
- |
- |
PREDICTED: uncharacterized protein LOC103997323 [Musa acuminata subsp. malaccensis] |
| 3 |
Hb_002027_260 |
0.0809507842 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 4 |
Hb_001141_330 |
0.081818903 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14850 [Jatropha curcas] |
| 5 |
Hb_000953_010 |
0.0834507845 |
- |
- |
hypothetical protein CISIN_1g0425012mg, partial [Citrus sinensis] |
| 6 |
Hb_004324_280 |
0.0852466762 |
- |
- |
PREDICTED: plastid division protein PDV1 [Jatropha curcas] |
| 7 |
Hb_000803_190 |
0.0872768845 |
- |
- |
PREDICTED: THUMP domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_156850_030 |
0.0877587026 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 9 |
Hb_000009_550 |
0.0877674303 |
- |
- |
hypothetical protein RCOM_0603640 [Ricinus communis] |
| 10 |
Hb_004734_060 |
0.088764493 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000787_110 |
0.0925256883 |
- |
- |
eukaryotic translation initiation factor 2 [Hevea brasiliensis] |
| 12 |
Hb_000352_090 |
0.0935534005 |
- |
- |
PREDICTED: syntaxin-22-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000836_360 |
0.0940578121 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000977_070 |
0.0949961547 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002154_030 |
0.0954124965 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_004934_090 |
0.0955757626 |
- |
- |
hypothetical protein JCGZ_08073 [Jatropha curcas] |
| 17 |
Hb_000162_090 |
0.0960275495 |
- |
- |
PREDICTED: methyltransferase-like protein 16 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004204_080 |
0.0962957252 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |
| 19 |
Hb_000923_020 |
0.0964061115 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 65-like [Jatropha curcas] |
| 20 |
Hb_001728_100 |
0.096441129 |
- |
- |
hypothetical protein JCGZ_03429 [Jatropha curcas] |