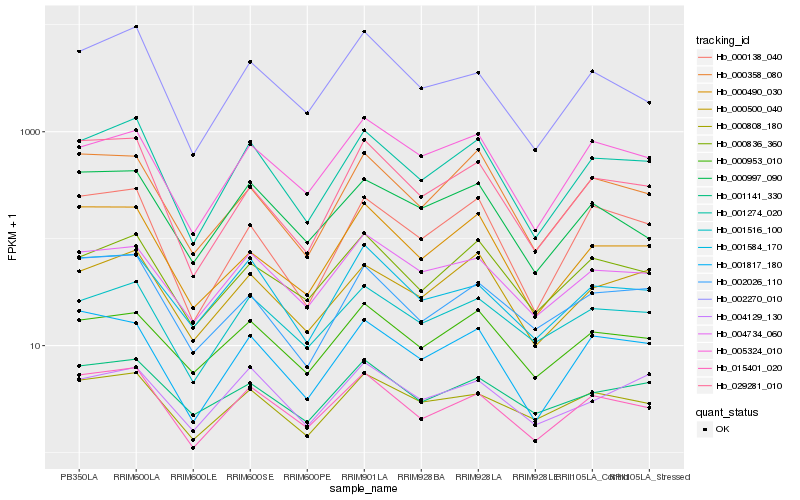
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001274_020 |
0.0 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter GCN1 [Hevea brasiliensis] |
| 2 |
Hb_015401_020 |
0.0898504826 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_000500_040 |
0.0912861939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000138_040 |
0.09312788 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 5 |
Hb_001516_100 |
0.0937941455 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000836_360 |
0.0975959707 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000808_180 |
0.0985226262 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09190 [Jatropha curcas] |
| 8 |
Hb_000953_010 |
0.1037627398 |
- |
- |
hypothetical protein CISIN_1g0425012mg, partial [Citrus sinensis] |
| 9 |
Hb_001141_330 |
0.1058210289 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14850 [Jatropha curcas] |
| 10 |
Hb_005324_010 |
0.1066380449 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 11 |
Hb_001817_180 |
0.1066788038 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 12 |
Hb_002270_010 |
0.108691756 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 13 |
Hb_000358_080 |
0.1093332015 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 14 |
Hb_004129_130 |
0.1096027694 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_029281_010 |
0.1097865951 |
- |
- |
hypothetical protein JCGZ_04025 [Jatropha curcas] |
| 16 |
Hb_004734_060 |
0.1106977152 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001584_170 |
0.1120070156 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 18 |
Hb_000997_090 |
0.1125584217 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 19 |
Hb_000490_030 |
0.1130435111 |
- |
- |
DNA-directed RNA polymerase II subunit, putative [Ricinus communis] |
| 20 |
Hb_002026_110 |
0.1135924024 |
- |
- |
protein binding protein, putative [Ricinus communis] |