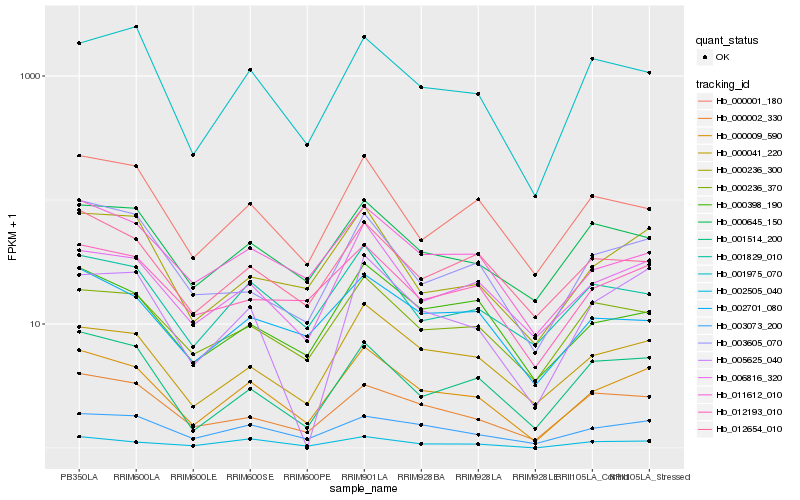
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_590 |
0.0 |
- |
- |
PREDICTED: prolyl endopeptidase-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003073_200 |
0.1085649709 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 3 |
Hb_000041_220 |
0.1090934278 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 4 |
Hb_006816_320 |
0.1166750914 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Jatropha curcas] |
| 5 |
Hb_000236_300 |
0.1184132978 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Jatropha curcas] |
| 6 |
Hb_002505_040 |
0.1191758345 |
- |
- |
- |
| 7 |
Hb_000398_190 |
0.1220818822 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 8 |
Hb_011612_010 |
0.1240753037 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 9 |
Hb_001514_200 |
0.1241288378 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 10 |
Hb_001975_070 |
0.1291520346 |
- |
- |
eukaryotic translation elongation factor 1B alpha-subunit [Hevea brasiliensis] |
| 11 |
Hb_000002_330 |
0.1294435241 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 12 |
Hb_005625_040 |
0.1308140523 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 13 |
Hb_000236_370 |
0.1314336289 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 14 |
Hb_012193_010 |
0.1315453575 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_001829_010 |
0.1329833154 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_002701_080 |
0.1330014725 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 17 |
Hb_003605_070 |
0.1337194511 |
- |
- |
PREDICTED: angio-associated migratory cell protein [Jatropha curcas] |
| 18 |
Hb_000001_180 |
0.1357022988 |
- |
- |
PREDICTED: uncharacterized protein LOC105648978 [Jatropha curcas] |
| 19 |
Hb_012654_010 |
0.1364554433 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 20 |
Hb_000645_150 |
0.1369637076 |
- |
- |
zinc finger protein, putative [Ricinus communis] |