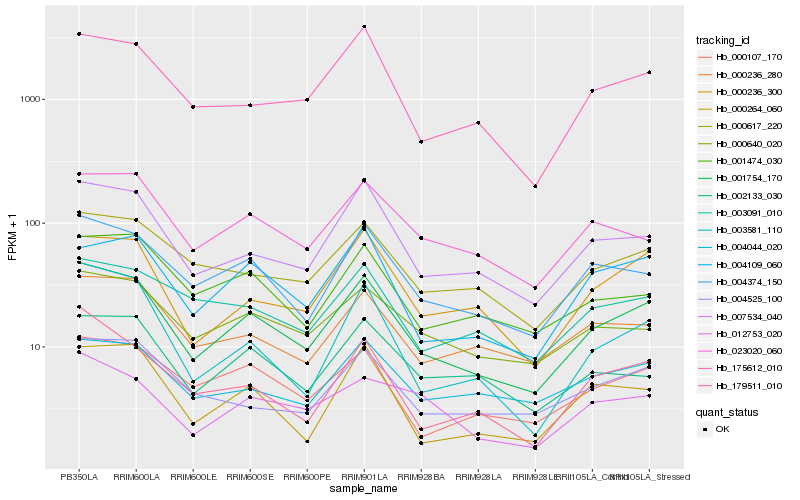
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_170 |
0.0 |
- |
- |
PREDICTED: 60S ribosome subunit biogenesis protein NIP7 homolog [Jatropha curcas] |
| 2 |
Hb_007534_040 |
0.0906530015 |
- |
- |
PREDICTED: uncharacterized protein LOC105635538 [Jatropha curcas] |
| 3 |
Hb_004374_150 |
0.0932803329 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000236_300 |
0.1027671676 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Jatropha curcas] |
| 5 |
Hb_175612_010 |
0.1046258207 |
transcription factor |
TF Family: MBF1 |
orf [Ricinus communis] |
| 6 |
Hb_000107_170 |
0.1090747514 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 7 |
Hb_000617_220 |
0.1115172661 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 8 |
Hb_004109_060 |
0.1138404215 |
- |
- |
hypothetical protein POPTR_0019s12520g [Populus trichocarpa] |
| 9 |
Hb_000640_020 |
0.1142589377 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 10 |
Hb_023020_060 |
0.1158839277 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 11 |
Hb_000236_280 |
0.1165597495 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_004525_100 |
0.1182485737 |
- |
- |
hypothetical protein JCGZ_18418 [Jatropha curcas] |
| 13 |
Hb_001474_030 |
0.1202692221 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 14 |
Hb_003581_110 |
0.120368731 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002133_030 |
0.1209153393 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 16 |
Hb_000264_060 |
0.1224535362 |
- |
- |
PREDICTED: RNA polymerase-associated protein RTF1 homolog [Jatropha curcas] |
| 17 |
Hb_012753_020 |
0.1225042926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004044_020 |
0.1229445306 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 19 |
Hb_179511_010 |
0.1233879431 |
- |
- |
- |
| 20 |
Hb_003091_010 |
0.1244401724 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |