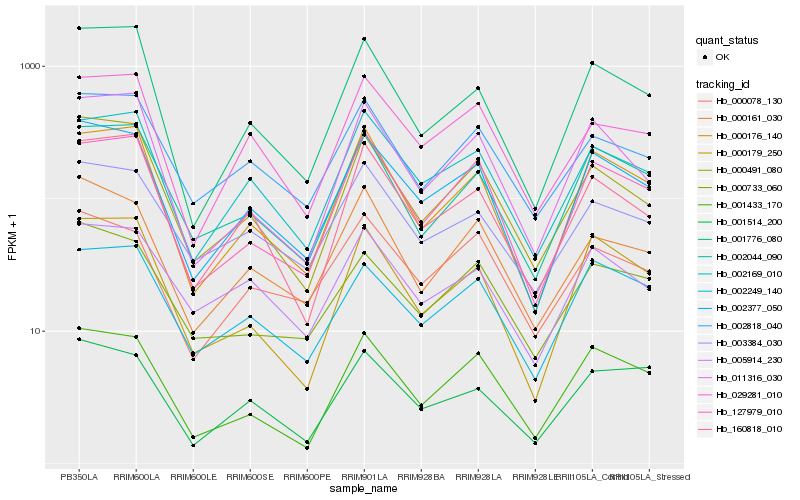
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002377_050 |
0.0 |
- |
- |
Agmatine deiminase, putative [Ricinus communis] |
| 2 |
Hb_002169_010 |
0.075880305 |
- |
- |
PREDICTED: la-related protein 6B [Jatropha curcas] |
| 3 |
Hb_000176_140 |
0.0764318177 |
- |
- |
PREDICTED: bystin [Jatropha curcas] |
| 4 |
Hb_001776_080 |
0.0816576297 |
- |
- |
PREDICTED: CAX-interacting protein 4 [Jatropha curcas] |
| 5 |
Hb_000491_080 |
0.0832216435 |
- |
- |
PREDICTED: F-box protein SKIP31 [Jatropha curcas] |
| 6 |
Hb_001433_170 |
0.0843422331 |
- |
- |
PREDICTED: topless-related protein 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_127979_010 |
0.0865865267 |
- |
- |
PREDICTED: uncharacterized protein LOC105629383 [Jatropha curcas] |
| 8 |
Hb_002249_140 |
0.0871773808 |
- |
- |
PREDICTED: uncharacterized protein LOC105648715 [Jatropha curcas] |
| 9 |
Hb_002044_090 |
0.087624909 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000179_250 |
0.0876292631 |
- |
- |
PREDICTED: WD repeat-containing protein 61-like [Citrus sinensis] |
| 11 |
Hb_000161_030 |
0.091322788 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP8 [Jatropha curcas] |
| 12 |
Hb_160818_010 |
0.091899679 |
- |
- |
- |
| 13 |
Hb_029281_010 |
0.0919841492 |
- |
- |
hypothetical protein JCGZ_04025 [Jatropha curcas] |
| 14 |
Hb_011316_030 |
0.0924922535 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005914_230 |
0.0942144474 |
- |
- |
PREDICTED: pre-mRNA-processing factor 19 homolog 1 [Jatropha curcas] |
| 16 |
Hb_000078_130 |
0.0949923831 |
- |
- |
- |
| 17 |
Hb_001514_200 |
0.0955485011 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 18 |
Hb_000733_060 |
0.0957332574 |
- |
- |
PREDICTED: protein GUCD1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002818_040 |
0.098151736 |
- |
- |
tRNA synthetase-related family protein [Populus trichocarpa] |
| 20 |
Hb_003384_030 |
0.0993217901 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit beta-like [Jatropha curcas] |