| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
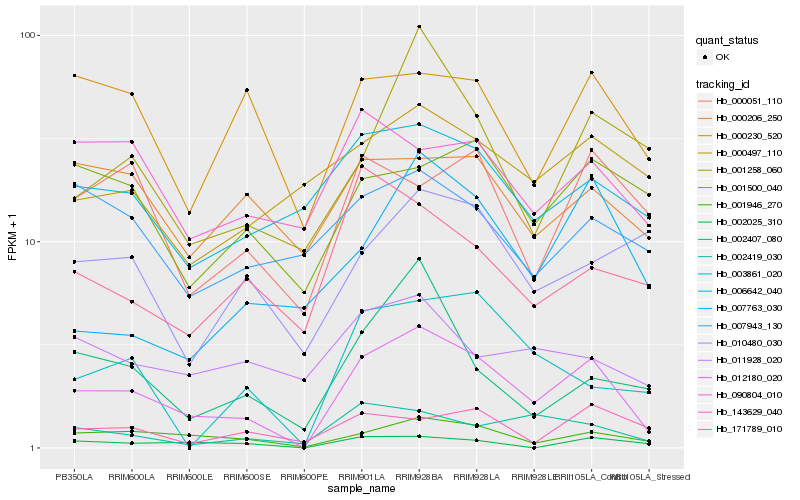
Hb_012180_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s15100g [Populus trichocarpa] |
| 2 |
Hb_001946_270 |
0.1721448913 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 3 |
Hb_002419_030 |
0.2274228009 |
- |
- |
- |
| 4 |
Hb_006642_040 |
0.2305045409 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 5 |
Hb_001258_060 |
0.2305689607 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 6 |
Hb_002407_080 |
0.2355051591 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 7 |
Hb_003861_020 |
0.2365436245 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like isoform X2 [Nelumbo nucifera] |
| 8 |
Hb_000230_520 |
0.2381243101 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 9 |
Hb_002025_310 |
0.2389684367 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000051_110 |
0.2394844661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007763_030 |
0.2410127592 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 12 |
Hb_001500_040 |
0.2437155722 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_011928_020 |
0.2470942877 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |
| 14 |
Hb_000206_250 |
0.2475342532 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 15 |
Hb_090804_010 |
0.247601934 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_143629_040 |
0.247760634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_171789_010 |
0.2491263131 |
- |
- |
phosphoenolpyruvate carboxylase [Glycine max] |
| 18 |
Hb_000497_110 |
0.2503113079 |
- |
- |
PREDICTED: uncharacterized protein LOC105647268 [Jatropha curcas] |
| 19 |
Hb_010480_030 |
0.2541695892 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_007943_130 |
0.2545490021 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |