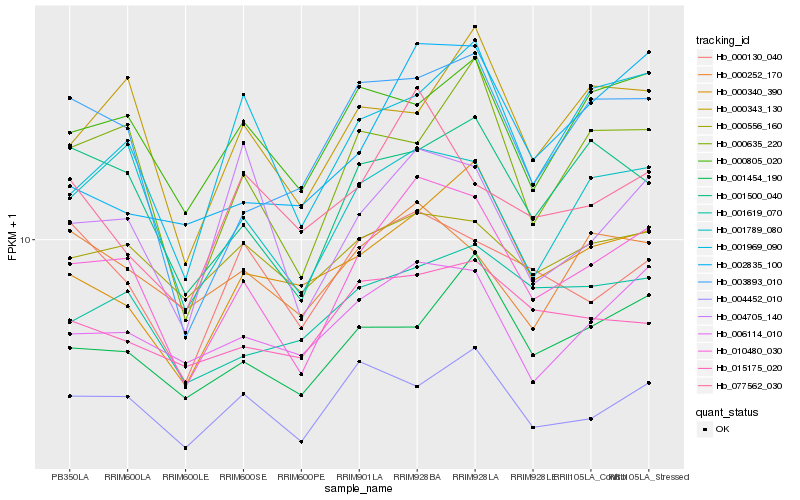
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010480_030 |
0.0 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_001789_080 |
0.1224839902 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 3 |
Hb_006114_010 |
0.127278769 |
- |
- |
PREDICTED: uncharacterized protein LOC105629068 [Jatropha curcas] |
| 4 |
Hb_001500_040 |
0.1356370291 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-1, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_003893_010 |
0.1360546246 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 6 |
Hb_000340_390 |
0.1436195098 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 7 |
Hb_000635_220 |
0.1438677799 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 8 |
Hb_001619_070 |
0.1447320905 |
- |
- |
PREDICTED: uncharacterized protein LOC103503585 [Cucumis melo] |
| 9 |
Hb_004452_010 |
0.146409122 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 10 |
Hb_004705_140 |
0.1464952577 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 11 |
Hb_000805_020 |
0.1466420988 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_001454_190 |
0.1498449737 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |
| 13 |
Hb_000556_160 |
0.1510604139 |
- |
- |
PREDICTED: ankyrin repeat and zinc finger domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_001969_090 |
0.1516433163 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 15 |
Hb_000130_040 |
0.1519076715 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 16 |
Hb_077562_030 |
0.1534370963 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_015175_020 |
0.1534816625 |
- |
- |
hypothetical protein JCGZ_14452 [Jatropha curcas] |
| 18 |
Hb_000252_170 |
0.1557592429 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000343_130 |
0.1562244083 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 20 |
Hb_002835_100 |
0.1563045464 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |