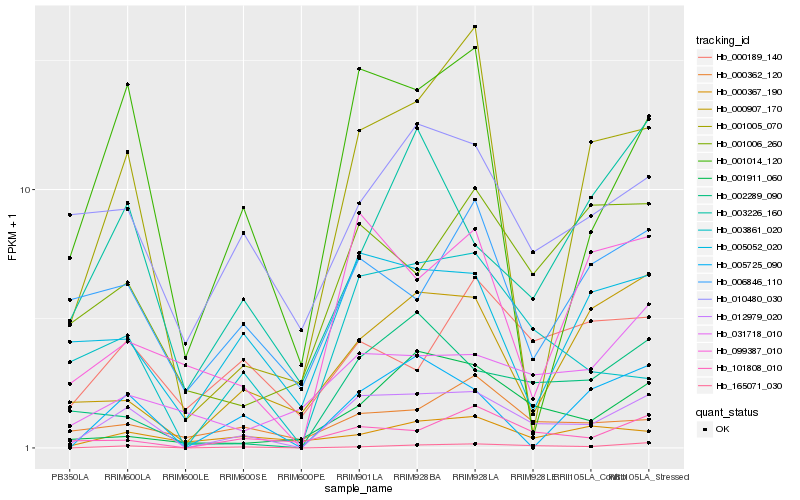
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012979_020 |
0.0 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Solanum lycopersicum] |
| 2 |
Hb_165071_030 |
0.2014990053 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_003861_020 |
0.2311292494 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like isoform X2 [Nelumbo nucifera] |
| 4 |
Hb_001014_120 |
0.2346753503 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor ERF113 [Jatropha curcas] |
| 5 |
Hb_002289_090 |
0.236981235 |
- |
- |
cytoplasmic tRNA 2-thiolation protein 2-like [Scleropages formosus] |
| 6 |
Hb_101808_010 |
0.2388888371 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005725_090 |
0.2469113544 |
- |
- |
PREDICTED: phytosulfokines 3 [Cicer arietinum] |
| 8 |
Hb_005052_020 |
0.2515403511 |
- |
- |
- |
| 9 |
Hb_010480_030 |
0.2530418433 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_099387_010 |
0.2535946289 |
- |
- |
- |
| 11 |
Hb_001006_260 |
0.25591263 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 12 |
Hb_001005_070 |
0.2564783313 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 13 |
Hb_003226_160 |
0.2591040266 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2-like isoform X2 [Populus euphratica] |
| 14 |
Hb_001911_060 |
0.2599876073 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000907_170 |
0.2624882912 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_031718_010 |
0.2630585854 |
- |
- |
- |
| 17 |
Hb_000362_120 |
0.2647535755 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 18 |
Hb_000367_190 |
0.2664628017 |
- |
- |
hypothetical protein RCOM_1470550 [Ricinus communis] |
| 19 |
Hb_006846_110 |
0.2666414066 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61800 [Jatropha curcas] |
| 20 |
Hb_000189_140 |
0.2675407449 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |