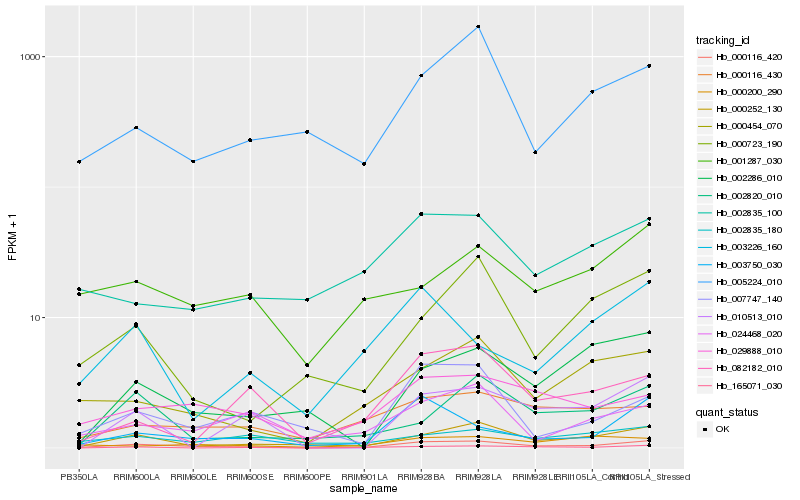
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_420 |
0.0 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2-like [Jatropha curcas] |
| 2 |
Hb_000252_130 |
0.175827449 |
- |
- |
hypothetical protein POPTR_0011s17240g, partial [Populus trichocarpa] |
| 3 |
Hb_000454_070 |
0.2103826627 |
- |
- |
PREDICTED: 30S ribosomal protein S31, mitochondrial [Cucumis sativus] |
| 4 |
Hb_029888_010 |
0.2191008187 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Nicotiana tomentosiformis] |
| 5 |
Hb_003750_030 |
0.2201568981 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 6 |
Hb_165071_030 |
0.2265163225 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_000116_430 |
0.2380031986 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002820_010 |
0.238924092 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005224_010 |
0.2401062627 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 10 |
Hb_000723_190 |
0.2416032417 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_010513_010 |
0.2420846966 |
- |
- |
- |
| 12 |
Hb_024468_020 |
0.2448079357 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 13 |
Hb_007747_140 |
0.2451419174 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002835_100 |
0.2466346631 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 15 |
Hb_001287_030 |
0.2469304558 |
- |
- |
30S ribosomal protein S31, mitochondrial precursor, putative [Ricinus communis] |
| 16 |
Hb_002286_010 |
0.2480591573 |
- |
- |
kinase-like protein pac.x.7.4, partial [Platanus x acerifolia] |
| 17 |
Hb_002835_180 |
0.2507722043 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_082182_010 |
0.2535711247 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 19 |
Hb_003226_160 |
0.2539816679 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2-like isoform X2 [Populus euphratica] |
| 20 |
Hb_000200_290 |
0.2542074487 |
- |
- |
PREDICTED: elongator complex protein 2 isoform X1 [Jatropha curcas] |