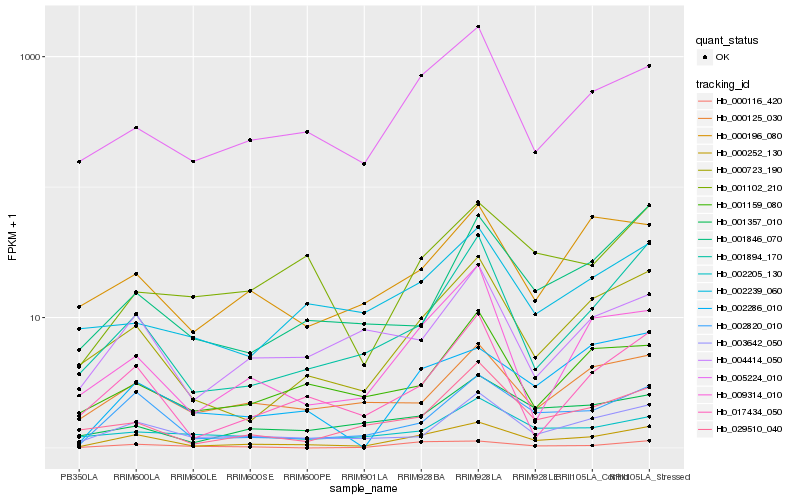
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_130 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s17240g, partial [Populus trichocarpa] |
| 2 |
Hb_002820_010 |
0.1238420367 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000723_190 |
0.1449074946 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_003642_050 |
0.1606952768 |
- |
- |
hypothetical protein CISIN_1g048193mg, partial [Citrus sinensis] |
| 5 |
Hb_005224_010 |
0.16850574 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 6 |
Hb_000116_420 |
0.175827449 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2-like [Jatropha curcas] |
| 7 |
Hb_001894_170 |
0.1784519849 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |
| 8 |
Hb_002286_010 |
0.1806739743 |
- |
- |
kinase-like protein pac.x.7.4, partial [Platanus x acerifolia] |
| 9 |
Hb_029510_040 |
0.1868556222 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 10 |
Hb_001357_010 |
0.1879263352 |
- |
- |
- |
| 11 |
Hb_000125_030 |
0.1882074087 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 12 |
Hb_017434_050 |
0.1911205537 |
- |
- |
PREDICTED: importin subunit beta-3-like [Jatropha curcas] |
| 13 |
Hb_004414_050 |
0.1920969931 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000196_080 |
0.1930403767 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001102_210 |
0.1979440728 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002239_060 |
0.1994516767 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 17 |
Hb_001846_070 |
0.1994713564 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 18 |
Hb_009314_010 |
0.1999337524 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 19 |
Hb_002205_130 |
0.2001867905 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 20 |
Hb_001159_080 |
0.2025786025 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |