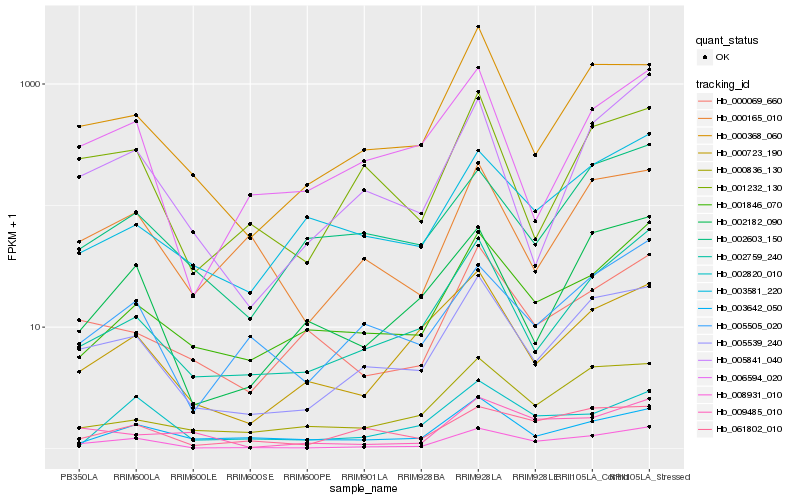
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008931_010 |
0.0 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 2 |
Hb_005539_240 |
0.1177495993 |
- |
- |
hypothetical protein MIMGU_mgv1a0220462mg, partial [Erythranthe guttata] |
| 3 |
Hb_005505_020 |
0.1572256834 |
- |
- |
Potassium channel AKT1, putative [Ricinus communis] |
| 4 |
Hb_002759_240 |
0.1659739061 |
- |
- |
hypothetical protein EUGRSUZ_E01319 [Eucalyptus grandis] |
| 5 |
Hb_001846_070 |
0.1684451454 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 6 |
Hb_061802_010 |
0.1725857362 |
- |
- |
hypothetical protein VITISV_011986 [Vitis vinifera] |
| 7 |
Hb_002182_090 |
0.1781047451 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF4.6-like isoform X1 [Gossypium raimondii] |
| 8 |
Hb_000069_660 |
0.1783943455 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000165_010 |
0.1786224706 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 10 |
Hb_000723_190 |
0.1802564036 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_009485_010 |
0.1866638873 |
- |
- |
PREDICTED: uncharacterized protein LOC105641071 [Jatropha curcas] |
| 12 |
Hb_000836_130 |
0.1866750252 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000368_060 |
0.1882231045 |
- |
- |
thioredoxin H-type 6 [Hevea brasiliensis] |
| 14 |
Hb_005841_040 |
0.1894544303 |
- |
- |
stress-induced hydrophobic peptide 1 [Hevea brasiliensis] |
| 15 |
Hb_001232_130 |
0.1899652908 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 16 |
Hb_003581_220 |
0.1905022274 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_006594_020 |
0.1925593541 |
- |
- |
PREDICTED: uncharacterized protein LOC105138974 isoform X1 [Populus euphratica] |
| 18 |
Hb_002603_150 |
0.1931780421 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 19 |
Hb_003642_050 |
0.1935358499 |
- |
- |
hypothetical protein CISIN_1g048193mg, partial [Citrus sinensis] |
| 20 |
Hb_002820_010 |
0.1955216654 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |