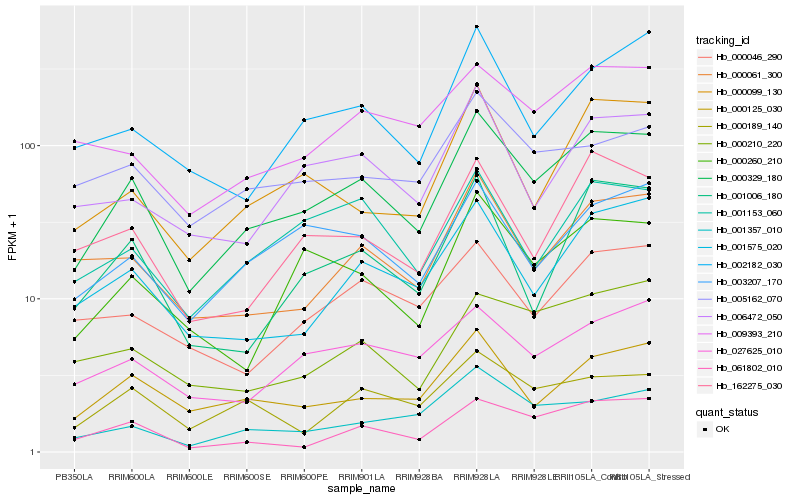
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_180 |
0.0 |
- |
- |
Auxin-induced protein X10A, putative [Ricinus communis] |
| 2 |
Hb_000125_030 |
0.1230959702 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 3 |
Hb_000260_210 |
0.1245266894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003207_170 |
0.1263879614 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000061_300 |
0.1275291205 |
- |
- |
hypothetical protein JCGZ_11668 [Jatropha curcas] |
| 6 |
Hb_061802_010 |
0.1277662879 |
- |
- |
hypothetical protein VITISV_011986 [Vitis vinifera] |
| 7 |
Hb_001575_020 |
0.1284251001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001153_060 |
0.1301806382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_162275_030 |
0.1316982474 |
- |
- |
PREDICTED: uncharacterized protein LOC105646638 [Jatropha curcas] |
| 10 |
Hb_027625_010 |
0.1320347595 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 11 |
Hb_006472_050 |
0.133128238 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 isoform X2 [Elaeis guineensis] |
| 12 |
Hb_000189_140 |
0.134446069 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_009393_210 |
0.1349235289 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 14 |
Hb_001357_010 |
0.1361755927 |
- |
- |
- |
| 15 |
Hb_000099_130 |
0.1379051865 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 16 |
Hb_001006_180 |
0.1386733384 |
- |
- |
PREDICTED: endonuclease V-like [Jatropha curcas] |
| 17 |
Hb_002182_030 |
0.1400252754 |
- |
- |
hypothetical protein Csa_5G139175 [Cucumis sativus] |
| 18 |
Hb_000210_220 |
0.1411731632 |
- |
- |
hypothetical protein POPTR_0006s22010g [Populus trichocarpa] |
| 19 |
Hb_000046_290 |
0.1418670228 |
- |
- |
PREDICTED: tubby-like F-box protein 3 [Tarenaya hassleriana] |
| 20 |
Hb_005162_070 |
0.1428795073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |