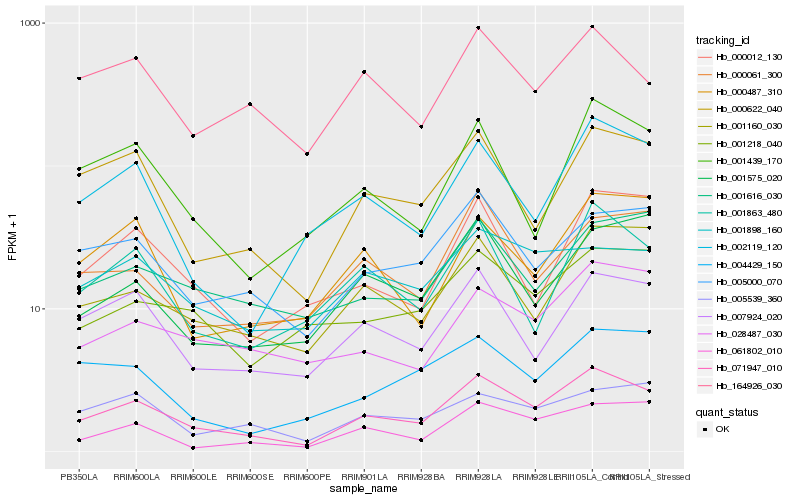
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071947_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000622_040 |
0.126410549 |
- |
- |
PREDICTED: uncharacterized protein LOC104596457 isoform X1 [Nelumbo nucifera] |
| 3 |
Hb_005000_070 |
0.1278537279 |
- |
- |
hypothetical protein POPTR_0012s01650g [Populus trichocarpa] |
| 4 |
Hb_007924_020 |
0.1310424533 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_001160_030 |
0.1314086433 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein POPTR_0018s05470g [Populus trichocarpa] |
| 6 |
Hb_061802_010 |
0.1325758011 |
- |
- |
hypothetical protein VITISV_011986 [Vitis vinifera] |
| 7 |
Hb_001863_480 |
0.1431859986 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 8 |
Hb_028487_030 |
0.1447493607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_164926_030 |
0.1450748831 |
- |
- |
PREDICTED: uncharacterized protein LOC105645602 [Jatropha curcas] |
| 10 |
Hb_002119_120 |
0.1456204193 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_001218_040 |
0.1463696444 |
- |
- |
Mitochondrial substrate carrier family protein [Theobroma cacao] |
| 12 |
Hb_000061_300 |
0.1466933427 |
- |
- |
hypothetical protein JCGZ_11668 [Jatropha curcas] |
| 13 |
Hb_001439_170 |
0.1476336385 |
- |
- |
PREDICTED: pectinesterase 31-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000012_130 |
0.1487741706 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 15 |
Hb_001616_030 |
0.1489951244 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor-like protein At4g13040 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005539_360 |
0.1498900359 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 17 |
Hb_001898_160 |
0.1503256121 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 18 |
Hb_001575_020 |
0.1510582624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004429_150 |
0.1514538352 |
- |
- |
- |
| 20 |
Hb_000487_310 |
0.1527821897 |
- |
- |
PREDICTED: uncharacterized protein LOC105646211 isoform X1 [Jatropha curcas] |