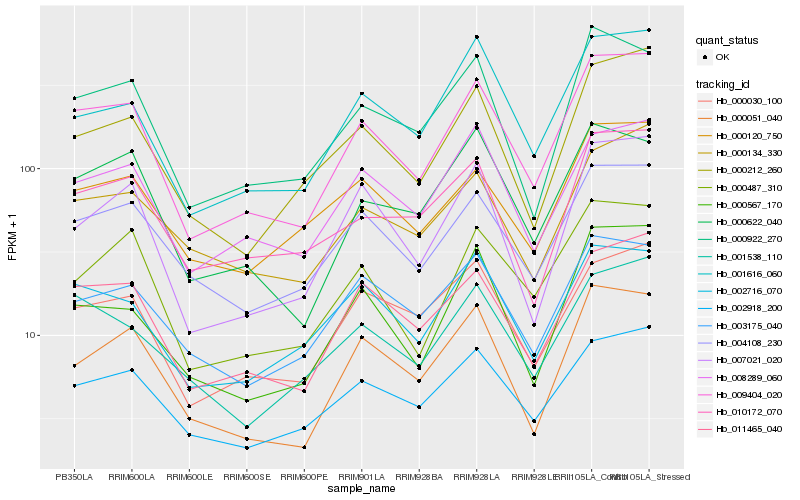
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009404_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011465_040 |
0.0690610631 |
- |
- |
PREDICTED: transcription and mRNA export factor SUS1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001616_060 |
0.0867140667 |
- |
- |
PREDICTED: DNA-directed RNA polymerase subunit 10-like protein [Jatropha curcas] |
| 4 |
Hb_000487_310 |
0.0868445963 |
- |
- |
PREDICTED: uncharacterized protein LOC105646211 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004108_230 |
0.0870082046 |
- |
- |
PREDICTED: 6.7 kDa chloroplast outer envelope membrane protein-like [Jatropha curcas] |
| 6 |
Hb_000922_270 |
0.08709053 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 7 |
Hb_002918_200 |
0.0878261919 |
- |
- |
- |
| 8 |
Hb_000212_260 |
0.0891150759 |
- |
- |
acyl-carrier-protein [Triadica sebifera] |
| 9 |
Hb_000051_040 |
0.0894587089 |
- |
- |
PREDICTED: uncharacterized protein LOC102597788 [Solanum tuberosum] |
| 10 |
Hb_007021_020 |
0.0895103641 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008289_060 |
0.0899566336 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000567_170 |
0.0921818971 |
- |
- |
PREDICTED: putative lipase YDR444W isoform X1 [Jatropha curcas] |
| 13 |
Hb_000030_100 |
0.0957284845 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |
| 14 |
Hb_002716_070 |
0.0968539272 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 15 |
Hb_000622_040 |
0.0988043556 |
- |
- |
PREDICTED: uncharacterized protein LOC104596457 isoform X1 [Nelumbo nucifera] |
| 16 |
Hb_001538_110 |
0.0996562051 |
- |
- |
PREDICTED: histone deacetylase 6, partial [Oryzias latipes] |
| 17 |
Hb_000120_750 |
0.1005181421 |
desease resistance |
Gene Name: NTPase_1 |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000134_330 |
0.1011218112 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm7 [Elaeis guineensis] |
| 19 |
Hb_010172_070 |
0.1018811111 |
- |
- |
PREDICTED: ELL-associated factor 1 [Jatropha curcas] |
| 20 |
Hb_003175_040 |
0.1023351882 |
- |
- |
PREDICTED: katanin p60 ATPase-containing subunit A1 isoform X1 [Jatropha curcas] |