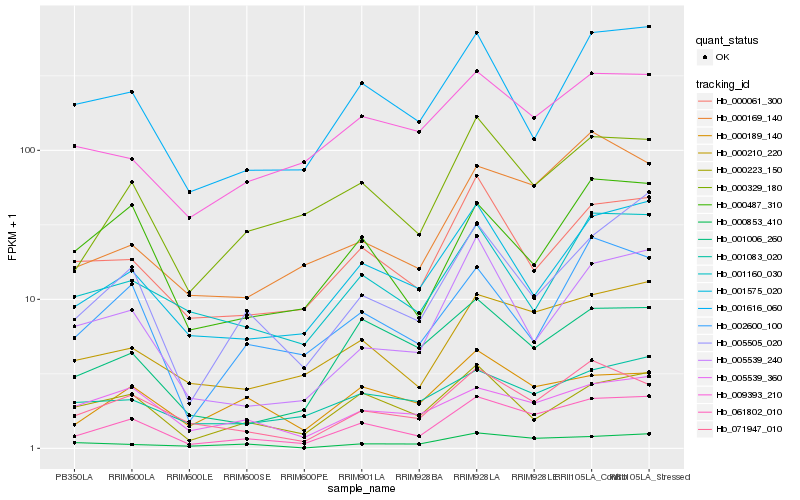
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061802_010 |
0.0 |
- |
- |
hypothetical protein VITISV_011986 [Vitis vinifera] |
| 2 |
Hb_000210_220 |
0.1161715657 |
- |
- |
hypothetical protein POPTR_0006s22010g [Populus trichocarpa] |
| 3 |
Hb_000329_180 |
0.1277662879 |
- |
- |
Auxin-induced protein X10A, putative [Ricinus communis] |
| 4 |
Hb_001575_020 |
0.1303434045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_071947_010 |
0.1325758011 |
- |
- |
- |
| 6 |
Hb_005505_020 |
0.1425487209 |
- |
- |
Potassium channel AKT1, putative [Ricinus communis] |
| 7 |
Hb_000061_300 |
0.1450971185 |
- |
- |
hypothetical protein JCGZ_11668 [Jatropha curcas] |
| 8 |
Hb_001006_260 |
0.1463061918 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 9 |
Hb_001616_060 |
0.1469157123 |
- |
- |
PREDICTED: DNA-directed RNA polymerase subunit 10-like protein [Jatropha curcas] |
| 10 |
Hb_000487_310 |
0.1491026571 |
- |
- |
PREDICTED: uncharacterized protein LOC105646211 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000169_140 |
0.1505069567 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 12 |
Hb_000853_410 |
0.1516508795 |
- |
- |
- |
| 13 |
Hb_009393_210 |
0.1548025912 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 14 |
Hb_001083_020 |
0.1570385322 |
- |
- |
- |
| 15 |
Hb_000223_150 |
0.1575172748 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Populus euphratica] |
| 16 |
Hb_002600_100 |
0.1575730756 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 17 |
Hb_001160_030 |
0.1581767399 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein POPTR_0018s05470g [Populus trichocarpa] |
| 18 |
Hb_000189_140 |
0.1587608138 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_005539_360 |
0.161693677 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 20 |
Hb_005539_240 |
0.1632994169 |
- |
- |
hypothetical protein MIMGU_mgv1a0220462mg, partial [Erythranthe guttata] |