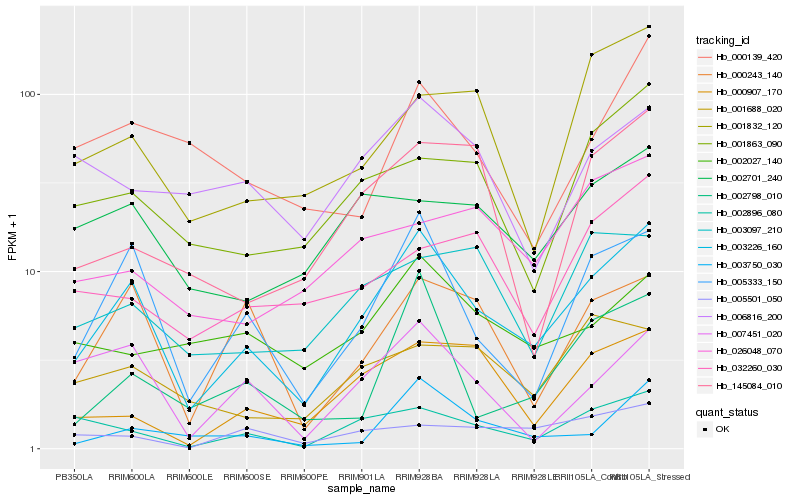
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_160 |
0.0 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2-like isoform X2 [Populus euphratica] |
| 2 |
Hb_005333_150 |
0.1405034182 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 3 |
Hb_002896_080 |
0.1955456902 |
- |
- |
- |
| 4 |
Hb_000243_140 |
0.1983253142 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 5 |
Hb_001863_090 |
0.2062784192 |
- |
- |
hypothetical protein POPTR_0006s07000g [Populus trichocarpa] |
| 6 |
Hb_000907_170 |
0.2099260401 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002798_010 |
0.2103078304 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 8 |
Hb_007451_020 |
0.2103555851 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 9 |
Hb_002701_240 |
0.2110966192 |
- |
- |
PREDICTED: putative DNA helicase INO80 [Jatropha curcas] |
| 10 |
Hb_001832_120 |
0.2122417164 |
- |
- |
hypothetical protein PHAVU_003G136800g [Phaseolus vulgaris] |
| 11 |
Hb_003750_030 |
0.2148671674 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 12 |
Hb_003097_210 |
0.2174840244 |
- |
- |
PREDICTED: uncharacterized protein LOC105631194 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002027_140 |
0.2182306222 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 14 |
Hb_001688_020 |
0.2204952902 |
- |
- |
PREDICTED: uncharacterized protein LOC103438026 [Malus domestica] |
| 15 |
Hb_145084_010 |
0.2215784775 |
- |
- |
PREDICTED: uncharacterized protein LOC105646270 [Jatropha curcas] |
| 16 |
Hb_000139_420 |
0.2217984853 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 17 |
Hb_005501_050 |
0.2240477625 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_006816_200 |
0.2249493163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_032260_030 |
0.2265572509 |
- |
- |
hypothetical protein POPTR_0001s10500g [Populus trichocarpa] |
| 20 |
Hb_026048_070 |
0.22692513 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |