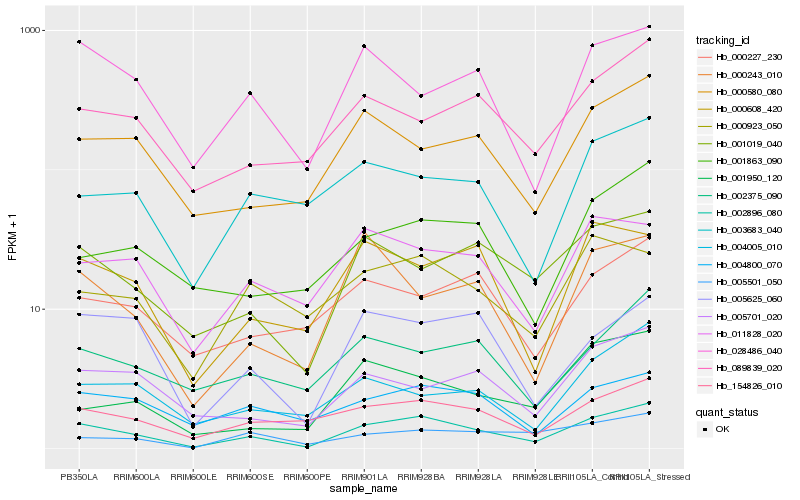
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002896_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_154826_010 |
0.1606452362 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_001019_040 |
0.1666897561 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005625_060 |
0.1756998252 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-1 [Jatropha curcas] |
| 5 |
Hb_000580_080 |
0.1765662916 |
- |
- |
hypothetical protein CICLE_v10006145mg [Citrus clementina] |
| 6 |
Hb_005701_020 |
0.1788322834 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 21 [Jatropha curcas] |
| 7 |
Hb_004800_070 |
0.179433645 |
- |
- |
- |
| 8 |
Hb_089839_020 |
0.1797183735 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 9 |
Hb_028486_040 |
0.1802358763 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 10 |
Hb_011828_020 |
0.180669044 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-10-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_001863_090 |
0.1815844033 |
- |
- |
hypothetical protein POPTR_0006s07000g [Populus trichocarpa] |
| 12 |
Hb_000608_420 |
0.1823297922 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 13 |
Hb_002375_090 |
0.1823498177 |
- |
- |
- |
| 14 |
Hb_003683_040 |
0.1847776119 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_005501_050 |
0.1849073101 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_004005_010 |
0.1871750664 |
- |
- |
PREDICTED: uncharacterized protein LOC102623333 isoform X2 [Citrus sinensis] |
| 17 |
Hb_000923_050 |
0.1874611425 |
- |
- |
PREDICTED: transcription factor bHLH113 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001950_120 |
0.1882729945 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000243_010 |
0.189187106 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 20 |
Hb_000227_230 |
0.1898243948 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |