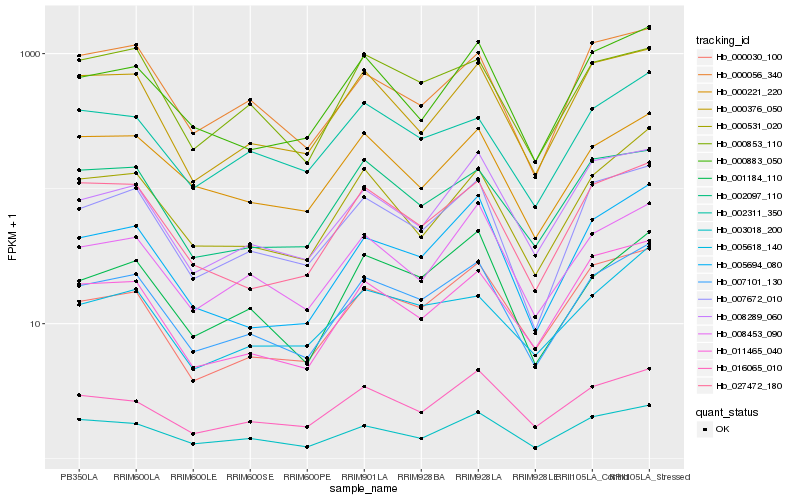
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007672_010 |
0.0611900441 |
- |
- |
PREDICTED: uncharacterized protein LOC105639669 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003018_200 |
0.0731144158 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 4 |
Hb_027472_180 |
0.0757099932 |
- |
- |
hypothetical protein L484_016229 [Morus notabilis] |
| 5 |
Hb_008453_090 |
0.076377333 |
- |
- |
hypothetical protein JCGZ_08177 [Jatropha curcas] |
| 6 |
Hb_001184_110 |
0.0786510428 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000376_050 |
0.0812537405 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000030_100 |
0.0819550506 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |
| 9 |
Hb_000883_050 |
0.0833492313 |
- |
- |
PREDICTED: 60S ribosomal protein L37a-like, partial [Solanum tuberosum] |
| 10 |
Hb_016065_010 |
0.0836139244 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08305 [Jatropha curcas] |
| 11 |
Hb_005694_080 |
0.0839605086 |
- |
- |
DNA-directed RNA polymerase III, putative [Ricinus communis] |
| 12 |
Hb_002097_110 |
0.0868306381 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000221_220 |
0.0872961618 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 14 |
Hb_002311_350 |
0.0879918569 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 15 |
Hb_000531_020 |
0.0881329617 |
- |
- |
PREDICTED: uncharacterized protein At4g28440-like [Populus euphratica] |
| 16 |
Hb_011465_040 |
0.0891444102 |
- |
- |
PREDICTED: transcription and mRNA export factor SUS1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005618_140 |
0.0894017371 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 18 |
Hb_008289_060 |
0.0895739176 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000853_110 |
0.0912819197 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 20 |
Hb_000056_340 |
0.0914679757 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |