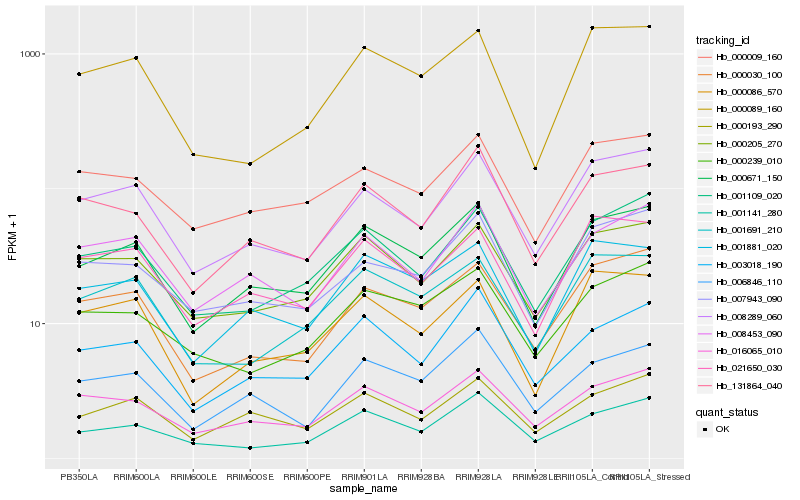
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000671_150 |
0.0 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 2 |
Hb_001881_020 |
0.0600758752 |
- |
- |
clathrin binding protein, putative [Ricinus communis] |
| 3 |
Hb_000205_270 |
0.0734022431 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000193_290 |
0.0770257049 |
- |
- |
hypothetical protein JCGZ_11661 [Jatropha curcas] |
| 5 |
Hb_001109_020 |
0.077136181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008289_060 |
0.0779941423 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003018_190 |
0.0791881379 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g31070, mitochondrial [Jatropha curcas] |
| 8 |
Hb_016065_010 |
0.0792390909 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08305 [Jatropha curcas] |
| 9 |
Hb_001691_210 |
0.0799401643 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 10 |
Hb_000030_100 |
0.0819869588 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |
| 11 |
Hb_006846_110 |
0.0825878783 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61800 [Jatropha curcas] |
| 12 |
Hb_131864_040 |
0.0827175635 |
- |
- |
PREDICTED: uncharacterized protein LOC105649406 isoform X1 [Jatropha curcas] |
| 13 |
Hb_021650_030 |
0.083645107 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 14 |
Hb_000086_570 |
0.0855171826 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 15 |
Hb_008453_090 |
0.0856616116 |
- |
- |
hypothetical protein JCGZ_08177 [Jatropha curcas] |
| 16 |
Hb_000009_160 |
0.0864862942 |
- |
- |
RUB1 conjugating enzyme 1 isoform 2 [Theobroma cacao] |
| 17 |
Hb_000089_160 |
0.0870356752 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 18 |
Hb_000239_010 |
0.0889475699 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_007943_090 |
0.0897727526 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 20 |
Hb_001141_280 |
0.089782474 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Vitis vinifera] |