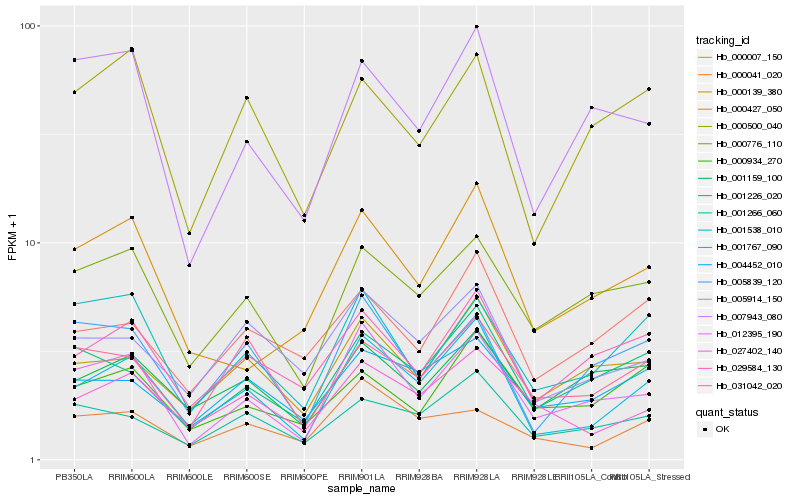
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001767_090 |
0.0 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 2 |
Hb_012395_190 |
0.1246173553 |
- |
- |
hypothetical protein B456_005G053800 [Gossypium raimondii] |
| 3 |
Hb_027402_140 |
0.1294468509 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g47530 [Jatropha curcas] |
| 4 |
Hb_000427_050 |
0.1370279552 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001266_060 |
0.1384446213 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g47840 [Jatropha curcas] |
| 6 |
Hb_005914_150 |
0.1405430783 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19890 [Jatropha curcas] |
| 7 |
Hb_029584_130 |
0.1407340429 |
- |
- |
PREDICTED: uncharacterized protein LOC105637865 [Jatropha curcas] |
| 8 |
Hb_001226_020 |
0.1481816354 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_004452_010 |
0.1483291664 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 10 |
Hb_000776_110 |
0.1518499194 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001159_100 |
0.1542707361 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g22760 [Jatropha curcas] |
| 12 |
Hb_005839_120 |
0.1551410402 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 13 |
Hb_000139_380 |
0.1556212238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000041_020 |
0.1578407234 |
- |
- |
hypothetical protein POPTR_0009s11640g [Populus trichocarpa] |
| 15 |
Hb_001538_010 |
0.1583459808 |
- |
- |
- |
| 16 |
Hb_031042_020 |
0.1584260225 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g47460 [Jatropha curcas] |
| 17 |
Hb_000007_150 |
0.1585196696 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41080 [Jatropha curcas] |
| 18 |
Hb_000500_040 |
0.1611211448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007943_080 |
0.1616952699 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000934_270 |
0.1621262413 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g21470 [Populus euphratica] |