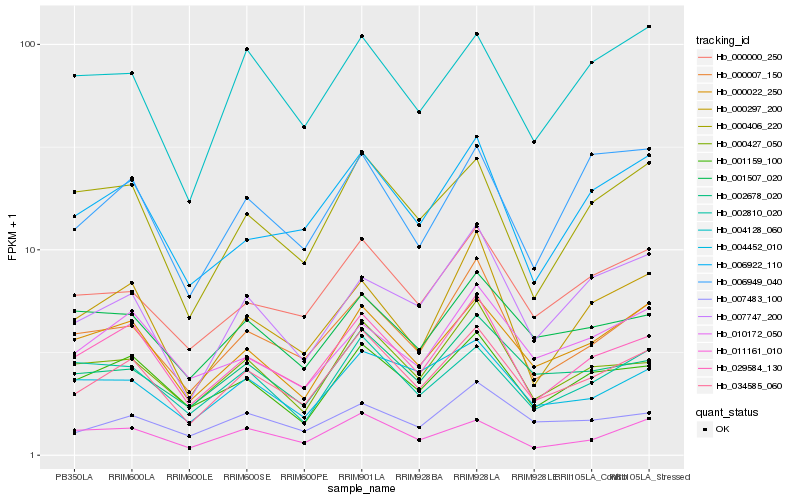
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034585_060 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g50420 [Jatropha curcas] |
| 2 |
Hb_029584_130 |
0.0711970589 |
- |
- |
PREDICTED: uncharacterized protein LOC105637865 [Jatropha curcas] |
| 3 |
Hb_000007_150 |
0.0817970516 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41080 [Jatropha curcas] |
| 4 |
Hb_000022_250 |
0.0822696649 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15130 [Jatropha curcas] |
| 5 |
Hb_000000_250 |
0.0834613576 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g19290 [Jatropha curcas] |
| 6 |
Hb_001159_100 |
0.0873654516 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g22760 [Jatropha curcas] |
| 7 |
Hb_002678_020 |
0.0895189769 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090-like [Jatropha curcas] |
| 8 |
Hb_006922_110 |
0.0923585665 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 9 |
Hb_000427_050 |
0.0940291259 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_007483_100 |
0.096262426 |
- |
- |
hypothetical protein POPTR_0001s34330g [Populus trichocarpa] |
| 11 |
Hb_000406_220 |
0.0979491079 |
- |
- |
hypothetical protein POPTR_0005s04700g [Populus trichocarpa] |
| 12 |
Hb_004452_010 |
0.1017589777 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 13 |
Hb_004128_060 |
0.1034798087 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit A [Jatropha curcas] |
| 14 |
Hb_006949_040 |
0.1037310022 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_011161_010 |
0.1039747568 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g35370 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000297_200 |
0.1051736076 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760-like [Vitis vinifera] |
| 17 |
Hb_002810_020 |
0.1056287289 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g25360 [Jatropha curcas] |
| 18 |
Hb_007747_200 |
0.1063256653 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g37570 [Jatropha curcas] |
| 19 |
Hb_001507_020 |
0.1090918437 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14170 [Jatropha curcas] |
| 20 |
Hb_010172_050 |
0.1092872692 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065 [Jatropha curcas] |