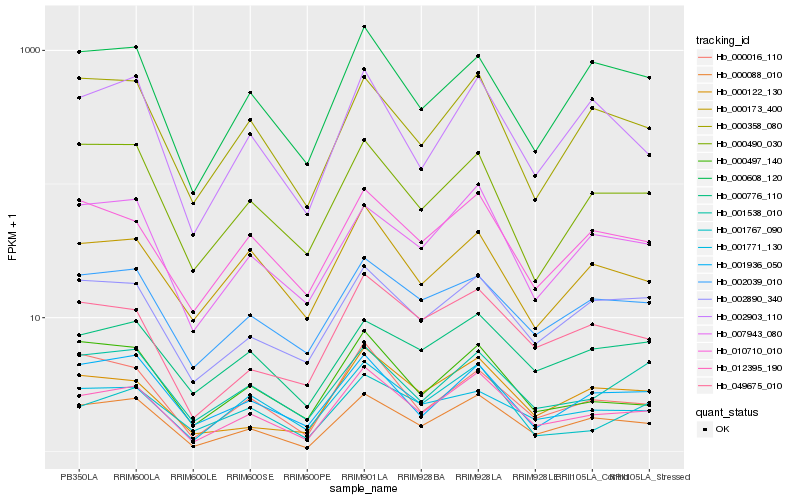
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_190 |
0.0 |
- |
- |
hypothetical protein B456_005G053800 [Gossypium raimondii] |
| 2 |
Hb_000088_010 |
0.0794388366 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_049675_010 |
0.1094097724 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 4 |
Hb_007943_080 |
0.1112632021 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000122_130 |
0.1124440469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010710_010 |
0.1128152495 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_000497_140 |
0.1155939843 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 8 |
Hb_002890_340 |
0.1168243377 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002039_010 |
0.1181535951 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 10 |
Hb_000358_080 |
0.1211469477 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 11 |
Hb_002903_110 |
0.121657296 |
- |
- |
PREDICTED: RNA polymerase-associated protein RTF1 homolog [Jatropha curcas] |
| 12 |
Hb_000173_400 |
0.1221242628 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18950 [Jatropha curcas] |
| 13 |
Hb_000776_110 |
0.1237662244 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001771_130 |
0.123855569 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g27610 [Jatropha curcas] |
| 15 |
Hb_000016_110 |
0.123992036 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 16 |
Hb_001767_090 |
0.1246173553 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 17 |
Hb_001538_010 |
0.1255511169 |
- |
- |
- |
| 18 |
Hb_001936_050 |
0.1258821213 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210-like [Prunus mume] |
| 19 |
Hb_000490_030 |
0.1262832593 |
- |
- |
DNA-directed RNA polymerase II subunit, putative [Ricinus communis] |
| 20 |
Hb_000608_120 |
0.127280884 |
- |
- |
PREDICTED: uncharacterized protein LOC105644872 [Jatropha curcas] |