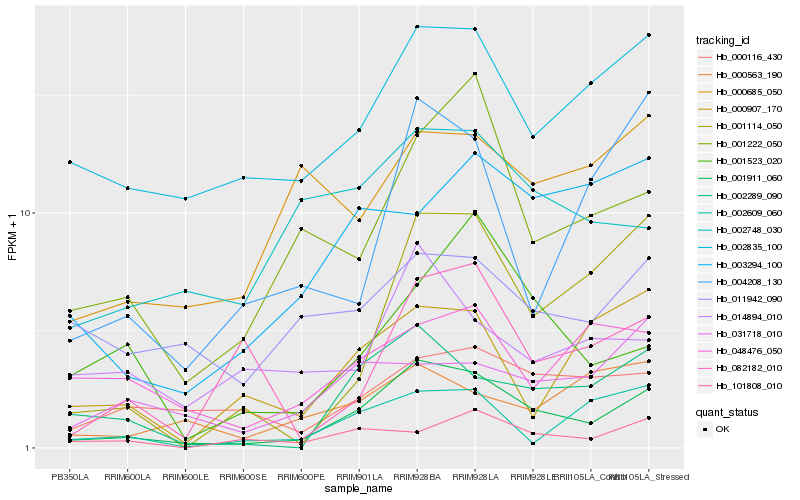
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001911_060 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002289_090 |
0.1935540485 |
- |
- |
cytoplasmic tRNA 2-thiolation protein 2-like [Scleropages formosus] |
| 3 |
Hb_002835_100 |
0.206257339 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 4 |
Hb_001114_050 |
0.2122112206 |
- |
- |
hypothetical protein RCOM_0744580 [Ricinus communis] |
| 5 |
Hb_001222_050 |
0.2165189155 |
- |
- |
hypothetical protein CICLE_v100161911mg, partial [Citrus clementina] |
| 6 |
Hb_002748_030 |
0.2310029493 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 7 |
Hb_000116_430 |
0.233364847 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003294_100 |
0.239783423 |
- |
- |
hypothetical protein VITISV_009629 [Vitis vinifera] |
| 9 |
Hb_004208_130 |
0.2414357475 |
- |
- |
hypothetical protein JCGZ_16532 [Jatropha curcas] |
| 10 |
Hb_002609_060 |
0.2422450762 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 11 |
Hb_082182_010 |
0.2447363102 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 12 |
Hb_000907_170 |
0.2473424813 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001523_020 |
0.2477295308 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 14 |
Hb_000685_050 |
0.2493244349 |
- |
- |
PREDICTED: mitochondrial inner membrane protein OXA1-like [Populus euphratica] |
| 15 |
Hb_011942_090 |
0.2499983165 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_101808_010 |
0.2503129892 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 17 |
Hb_048476_050 |
0.2526304526 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |
| 18 |
Hb_031718_010 |
0.2530128357 |
- |
- |
- |
| 19 |
Hb_000563_190 |
0.2531220918 |
- |
- |
hypothetical protein PRUPE_ppa002828mg [Prunus persica] |
| 20 |
Hb_014894_010 |
0.2547472185 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |