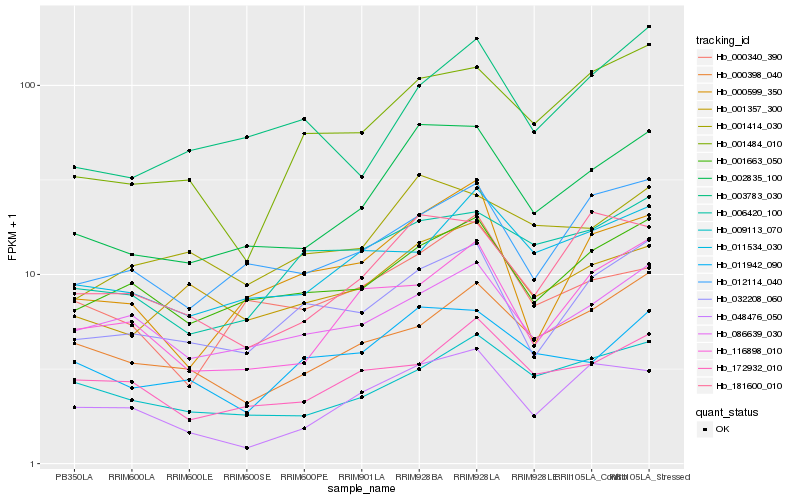
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_100 |
0.0 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 2 |
Hb_181600_010 |
0.1131565496 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 3 |
Hb_012114_040 |
0.1174662901 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_001663_050 |
0.1193734288 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 5 |
Hb_032208_060 |
0.1200069632 |
- |
- |
PREDICTED: constitutive photomorphogenesis protein 10 isoform X2 [Populus euphratica] |
| 6 |
Hb_001357_300 |
0.1212236806 |
- |
- |
PREDICTED: imidazole glycerol phosphate synthase hisHF, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_001414_030 |
0.1222824959 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 8 |
Hb_011942_090 |
0.1243253697 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001484_010 |
0.1289647744 |
- |
- |
hypothetical protein POPTR_0005s07440g [Populus trichocarpa] |
| 10 |
Hb_048476_050 |
0.1307669856 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |
| 11 |
Hb_000599_350 |
0.1310109604 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_009113_070 |
0.1310581403 |
- |
- |
hypothetical protein POPTR_0010s07510g [Populus trichocarpa] |
| 13 |
Hb_006420_100 |
0.1321938314 |
- |
- |
PREDICTED: protein bicaudal C homolog 1 [Jatropha curcas] |
| 14 |
Hb_116898_010 |
0.1324906018 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |
| 15 |
Hb_086639_030 |
0.1360255568 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 16 |
Hb_011534_030 |
0.1386499095 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 17 |
Hb_000398_040 |
0.1391867844 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 18 |
Hb_172932_010 |
0.1418780282 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000340_390 |
0.14211335 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 20 |
Hb_003783_030 |
0.1430225031 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |