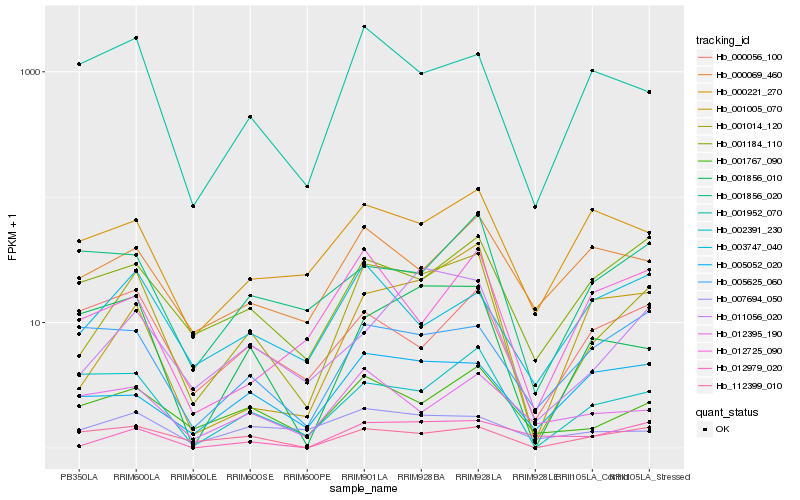
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001014_120 |
0.0 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor ERF113 [Jatropha curcas] |
| 2 |
Hb_001767_090 |
0.1878380285 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 3 |
Hb_011056_020 |
0.2030419027 |
transcription factor |
TF Family: ERF |
AP2 domain transcription factor RAP2.3, putative [Ricinus communis] |
| 4 |
Hb_001005_070 |
0.2116443822 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 5 |
Hb_001184_110 |
0.2217944575 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002391_230 |
0.2225749134 |
- |
- |
hypothetical protein RCOM_0745080 [Ricinus communis] |
| 7 |
Hb_112399_010 |
0.2253738664 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_001856_010 |
0.2261174115 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_005052_020 |
0.2278535671 |
- |
- |
- |
| 10 |
Hb_001952_070 |
0.2316987672 |
- |
- |
40S ribosomal protein S6B [Hevea brasiliensis] |
| 11 |
Hb_012979_020 |
0.2346753503 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Solanum lycopersicum] |
| 12 |
Hb_000221_270 |
0.2401195589 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 13 |
Hb_003747_040 |
0.2415800332 |
- |
- |
- |
| 14 |
Hb_000069_460 |
0.2429346264 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 15 |
Hb_005625_060 |
0.2432172319 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-1 [Jatropha curcas] |
| 16 |
Hb_007694_050 |
0.2445687125 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 17 |
Hb_012395_190 |
0.245421883 |
- |
- |
hypothetical protein B456_005G053800 [Gossypium raimondii] |
| 18 |
Hb_012725_090 |
0.2463097215 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 19 |
Hb_000056_100 |
0.2465541685 |
- |
- |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001856_020 |
0.2472400473 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |