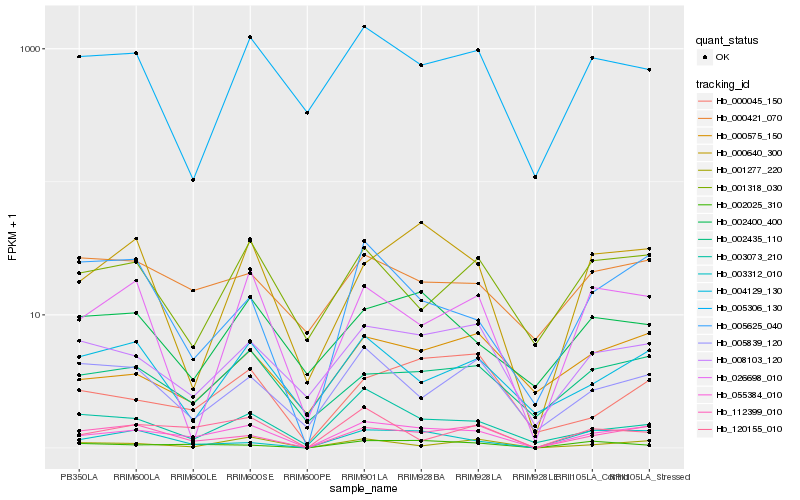
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055384_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_112399_010 |
0.14972257 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_002435_110 |
0.1885755519 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 4 |
Hb_008103_120 |
0.1895437176 |
- |
- |
hypothetical protein CISIN_1g009650mg [Citrus sinensis] |
| 5 |
Hb_120155_010 |
0.1911613463 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_002025_310 |
0.1997155956 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_003073_210 |
0.2002327757 |
- |
- |
hypothetical protein CISIN_1g036722mg, partial [Citrus sinensis] |
| 8 |
Hb_000640_300 |
0.2017961486 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 9 |
Hb_001277_220 |
0.2042112714 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 10 |
Hb_005625_040 |
0.2106881306 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 11 |
Hb_003312_010 |
0.210864878 |
- |
- |
PREDICTED: uncharacterized protein LOC104428797 [Eucalyptus grandis] |
| 12 |
Hb_000045_150 |
0.2120077356 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 13 |
Hb_000421_070 |
0.2120955989 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_005839_120 |
0.2162971622 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 15 |
Hb_000575_150 |
0.2163734274 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 16 |
Hb_002400_400 |
0.2183769395 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_004129_130 |
0.2185177881 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_026698_010 |
0.2200829929 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_001318_030 |
0.2201732749 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 20 |
Hb_005306_130 |
0.2203623016 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |