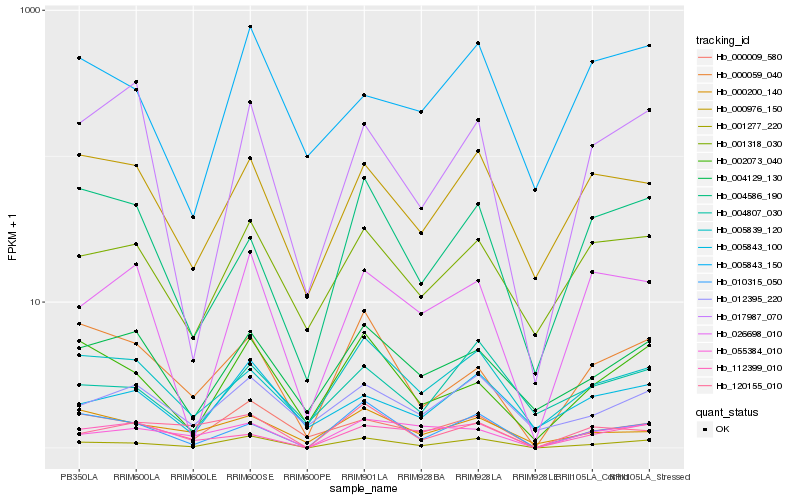
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_220 |
0.0 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 2 |
Hb_010315_050 |
0.1655456493 |
- |
- |
hypothetical protein JCGZ_25247 [Jatropha curcas] |
| 3 |
Hb_000009_580 |
0.1822172834 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 4 |
Hb_002073_040 |
0.1902650308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000059_040 |
0.1941304806 |
- |
- |
Uncharacterized protein TCM_020504 [Theobroma cacao] |
| 6 |
Hb_001318_030 |
0.2004616232 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 7 |
Hb_026698_010 |
0.2017944264 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 8 |
Hb_005839_120 |
0.2019789015 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 9 |
Hb_004129_130 |
0.2041685506 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_055384_010 |
0.2042112714 |
- |
- |
- |
| 11 |
Hb_000976_150 |
0.2045388274 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45 [Jatropha curcas] |
| 12 |
Hb_005843_100 |
0.2056582311 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350-like [Jatropha curcas] |
| 13 |
Hb_012395_220 |
0.206590201 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 14 |
Hb_004807_030 |
0.2091572466 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 15 |
Hb_120155_010 |
0.2160533461 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_017987_070 |
0.2167182522 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT22-like [Jatropha curcas] |
| 17 |
Hb_004586_190 |
0.2204458763 |
- |
- |
PREDICTED: protein CutA, chloroplastic [Jatropha curcas] |
| 18 |
Hb_112399_010 |
0.2238385283 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_005843_150 |
0.2238946848 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 20 |
Hb_000200_140 |
0.2242054621 |
- |
- |
PREDICTED: uncharacterized protein LOC104591989 [Nelumbo nucifera] |