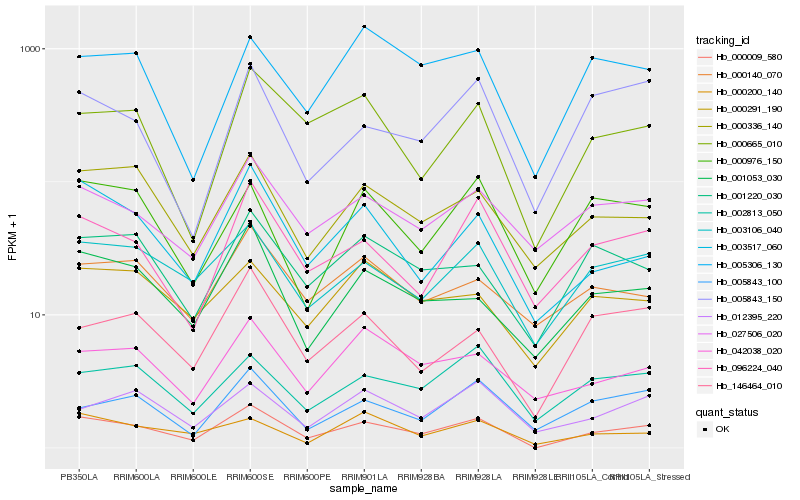
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_580 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 2 |
Hb_003106_040 |
0.140510698 |
- |
- |
lipase, putative [Ricinus communis] |
| 3 |
Hb_000665_010 |
0.147407413 |
- |
- |
hypothetical protein 20 [Hevea brasiliensis] |
| 4 |
Hb_003517_060 |
0.148847343 |
- |
- |
hypothetical protein CISIN_1g044999mg, partial [Citrus sinensis] |
| 5 |
Hb_096224_040 |
0.1491242373 |
- |
- |
PREDICTED: probable nucleoredoxin 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001053_030 |
0.1542374837 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000336_140 |
0.1543411372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_146464_010 |
0.1563244689 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_027506_020 |
0.1580024517 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 10 |
Hb_001220_030 |
0.1595412045 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 11 |
Hb_002813_050 |
0.1600342566 |
desease resistance |
Gene Name: TIR_2 |
PREDICTED: uncharacterized protein LOC105635699 [Jatropha curcas] |
| 12 |
Hb_042038_020 |
0.1679225322 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000291_190 |
0.1700551509 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 14 |
Hb_000976_150 |
0.1701374253 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45 [Jatropha curcas] |
| 15 |
Hb_005843_150 |
0.1709711849 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 16 |
Hb_005843_100 |
0.1719240008 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350-like [Jatropha curcas] |
| 17 |
Hb_000200_140 |
0.1728242275 |
- |
- |
PREDICTED: uncharacterized protein LOC104591989 [Nelumbo nucifera] |
| 18 |
Hb_000140_070 |
0.1730205829 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 19 |
Hb_012395_220 |
0.1732803565 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 20 |
Hb_005306_130 |
0.1734239097 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |