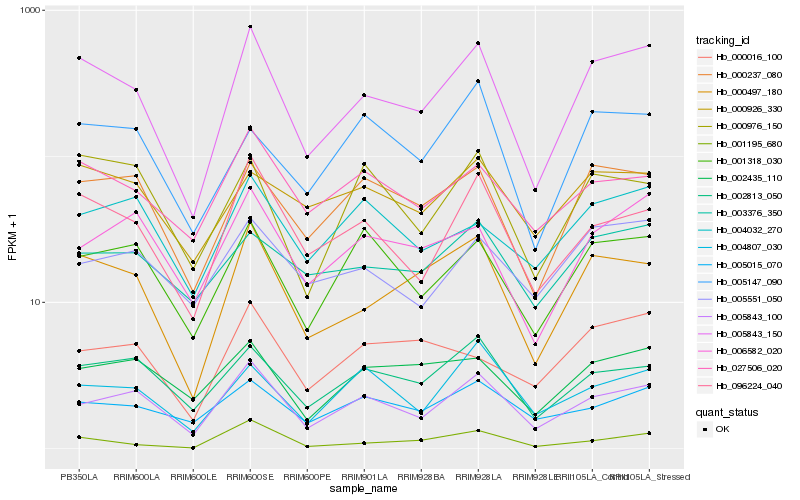
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005843_150 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 2 |
Hb_000497_180 |
0.0949788676 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 3 |
Hb_005843_100 |
0.1139016099 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350-like [Jatropha curcas] |
| 4 |
Hb_096224_040 |
0.1216563132 |
- |
- |
PREDICTED: probable nucleoredoxin 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000237_080 |
0.1302517339 |
transcription factor |
TF Family: PHD |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |
| 6 |
Hb_002813_050 |
0.1434564792 |
desease resistance |
Gene Name: TIR_2 |
PREDICTED: uncharacterized protein LOC105635699 [Jatropha curcas] |
| 7 |
Hb_001318_030 |
0.144895827 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 8 |
Hb_006582_020 |
0.1495277869 |
- |
- |
Zinc finger protein ZFPM1 [Theobroma cacao] |
| 9 |
Hb_027506_020 |
0.1526537533 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 10 |
Hb_002435_110 |
0.1562605907 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 11 |
Hb_001195_680 |
0.1573999777 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 12 |
Hb_000976_150 |
0.1577035663 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45 [Jatropha curcas] |
| 13 |
Hb_003376_350 |
0.1581276058 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005551_050 |
0.1582052677 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005015_070 |
0.1616242012 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 16 |
Hb_005147_090 |
0.1619777332 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004807_030 |
0.1623574138 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 18 |
Hb_000016_100 |
0.163779806 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP19 [Jatropha curcas] |
| 19 |
Hb_000926_330 |
0.1638034763 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 20 |
Hb_004032_270 |
0.1671395092 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |