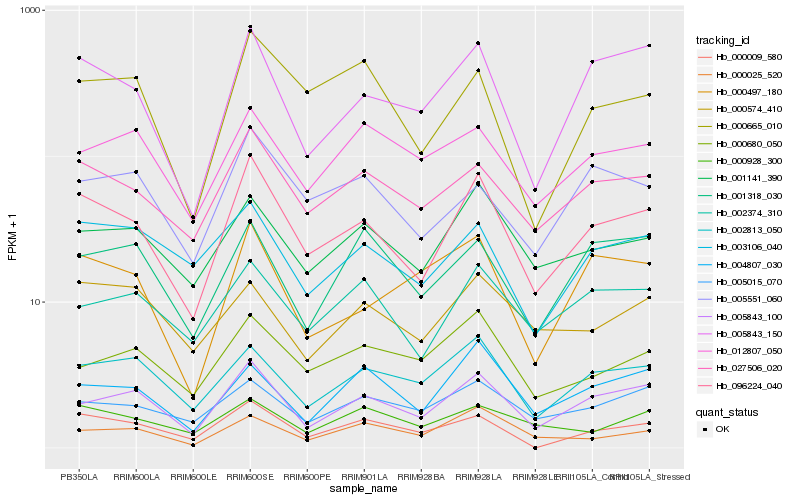
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096224_040 |
0.0 |
- |
- |
PREDICTED: probable nucleoredoxin 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005843_100 |
0.1106595992 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350-like [Jatropha curcas] |
| 3 |
Hb_027506_020 |
0.1155548883 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 4 |
Hb_005843_150 |
0.1216563132 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 5 |
Hb_001141_390 |
0.1331974979 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 6 |
Hb_002813_050 |
0.1388807545 |
desease resistance |
Gene Name: TIR_2 |
PREDICTED: uncharacterized protein LOC105635699 [Jatropha curcas] |
| 7 |
Hb_000665_010 |
0.1402789243 |
- |
- |
hypothetical protein 20 [Hevea brasiliensis] |
| 8 |
Hb_002374_310 |
0.144303632 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 9 |
Hb_003106_040 |
0.1465961205 |
- |
- |
lipase, putative [Ricinus communis] |
| 10 |
Hb_005551_060 |
0.1476052356 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 11 |
Hb_000009_580 |
0.1491242373 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 12 |
Hb_005015_070 |
0.1526905375 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 13 |
Hb_004807_030 |
0.153381671 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 14 |
Hb_000497_180 |
0.1535531331 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 15 |
Hb_000680_050 |
0.1548444069 |
- |
- |
hypothetical protein JCGZ_24767 [Jatropha curcas] |
| 16 |
Hb_000025_520 |
0.159695685 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27110 [Jatropha curcas] |
| 17 |
Hb_000574_410 |
0.1632793085 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600 [Jatropha curcas] |
| 18 |
Hb_012807_050 |
0.1634975204 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 19 |
Hb_000928_300 |
0.1639911544 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g03580 [Jatropha curcas] |
| 20 |
Hb_001318_030 |
0.1645687349 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |