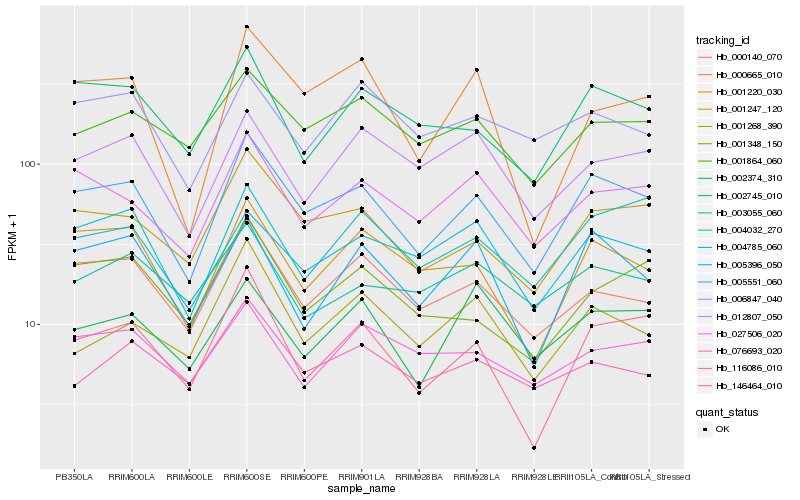
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005551_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 2 |
Hb_001247_120 |
0.0840987849 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_027506_020 |
0.1030251997 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 4 |
Hb_001220_030 |
0.1031886613 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 5 |
Hb_000140_070 |
0.103361409 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002745_010 |
0.1088460024 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 7 |
Hb_146464_010 |
0.1092009876 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004032_270 |
0.1104779783 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_001864_060 |
0.1165016344 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 10 |
Hb_004785_060 |
0.1180216477 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 11 |
Hb_005396_050 |
0.1201630224 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 12 |
Hb_001348_150 |
0.1248424881 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000665_010 |
0.1282753105 |
- |
- |
hypothetical protein 20 [Hevea brasiliensis] |
| 14 |
Hb_116086_010 |
0.1285863402 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 15 |
Hb_012807_050 |
0.1289989658 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 16 |
Hb_006847_040 |
0.1307926957 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 1 [Jatropha curcas] |
| 17 |
Hb_076693_020 |
0.1310602106 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 18 |
Hb_002374_310 |
0.1313050461 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 19 |
Hb_003055_060 |
0.1324572134 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001268_390 |
0.1328546926 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |