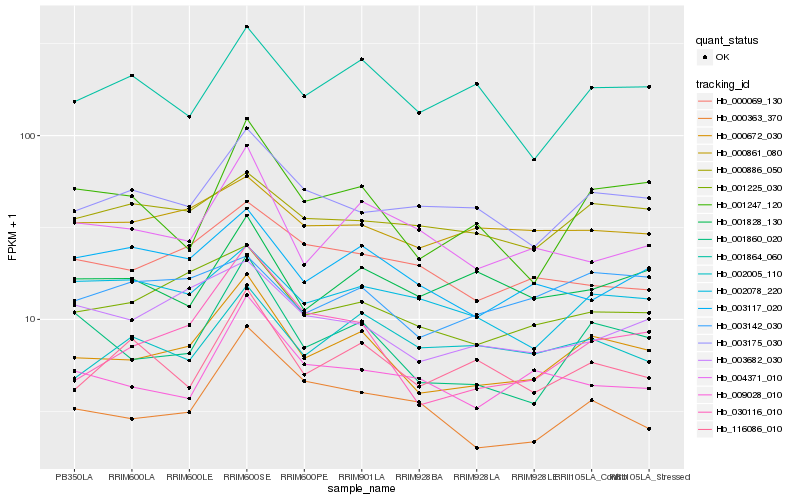
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000672_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001860_020 |
0.0909830361 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 3 |
Hb_001225_030 |
0.1000678083 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 4 |
Hb_030116_010 |
0.1085064337 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 5 |
Hb_001247_120 |
0.1105396004 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 6 |
Hb_003175_030 |
0.1121026386 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 7 |
Hb_003682_030 |
0.1165654307 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 8 |
Hb_116086_010 |
0.1198717748 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 9 |
Hb_004371_010 |
0.1200047283 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 10 |
Hb_001864_060 |
0.1212817067 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 11 |
Hb_003117_020 |
0.1229492147 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 12 |
Hb_009028_010 |
0.1231343433 |
- |
- |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 13 |
Hb_002078_220 |
0.1248122465 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001828_130 |
0.1248994015 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 15 |
Hb_000886_050 |
0.1270218696 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 16 |
Hb_000363_370 |
0.1274704792 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 17 |
Hb_002005_110 |
0.1276591662 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 18 |
Hb_003142_030 |
0.1277837194 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000861_080 |
0.1280455158 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 20 |
Hb_000069_130 |
0.1283131083 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |