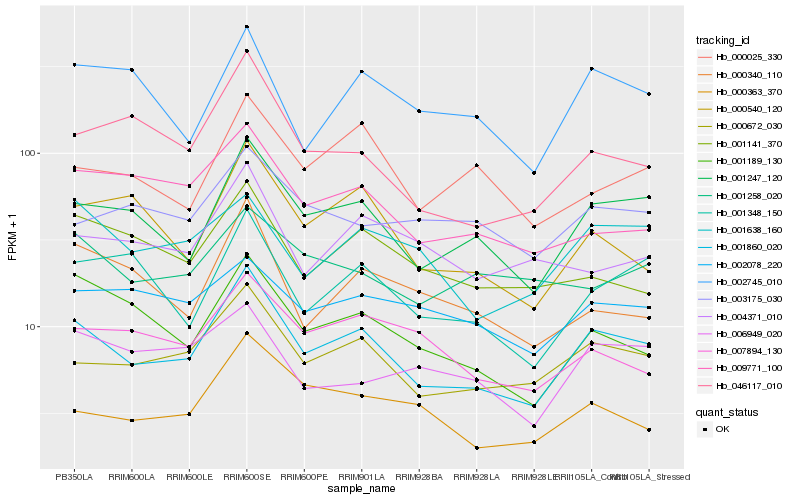
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001860_020 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 2 |
Hb_001247_120 |
0.0878322493 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_000672_030 |
0.0909830361 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001189_130 |
0.1246239257 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 5 |
Hb_046117_010 |
0.1259997817 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 6 |
Hb_001348_150 |
0.1287527435 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000540_120 |
0.1315881285 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 8 |
Hb_000363_370 |
0.1321894899 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 9 |
Hb_002745_010 |
0.1336279142 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 10 |
Hb_001638_160 |
0.1354472125 |
- |
- |
PREDICTED: uncharacterized protein LOC105642785 [Jatropha curcas] |
| 11 |
Hb_009771_100 |
0.1377050736 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 12 |
Hb_007894_130 |
0.1378221711 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 13 |
Hb_000025_330 |
0.1389190684 |
- |
- |
hypothetical protein JCGZ_22759 [Jatropha curcas] |
| 14 |
Hb_001141_370 |
0.1407884738 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 15 |
Hb_000340_110 |
0.1421042752 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 16 |
Hb_006949_020 |
0.1441190145 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine-C(5))-methyltransferase isoform X1 [Jatropha curcas] |
| 17 |
Hb_004371_010 |
0.1447729629 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 18 |
Hb_002078_220 |
0.1450698522 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001258_020 |
0.1472994653 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 20 |
Hb_003175_030 |
0.1475567272 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |