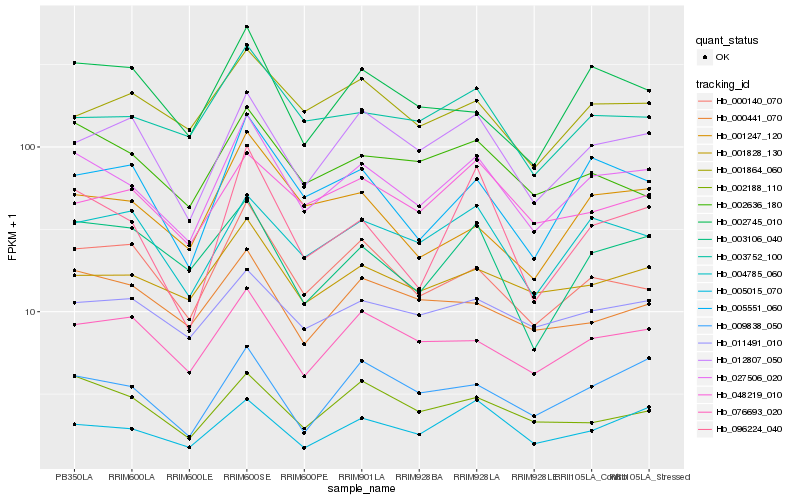
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027506_020 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 2 |
Hb_001828_130 |
0.0960748497 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 3 |
Hb_003752_100 |
0.0999985152 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 4 |
Hb_000140_070 |
0.1005758853 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 5 |
Hb_076693_020 |
0.1011695473 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 6 |
Hb_004785_060 |
0.1027081498 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 7 |
Hb_012807_050 |
0.1029767222 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 8 |
Hb_005551_060 |
0.1030251997 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 9 |
Hb_005015_070 |
0.10318593 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 10 |
Hb_011491_010 |
0.1062082635 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 11 |
Hb_002636_180 |
0.1078154659 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 12 |
Hb_003106_040 |
0.1106662293 |
- |
- |
lipase, putative [Ricinus communis] |
| 13 |
Hb_009838_050 |
0.1107098262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002188_110 |
0.1120440962 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 15 |
Hb_048219_010 |
0.1122019671 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 16 |
Hb_001864_060 |
0.113585008 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 17 |
Hb_000441_070 |
0.1143648615 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001247_120 |
0.1151098023 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_002745_010 |
0.1151207333 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 20 |
Hb_096224_040 |
0.1155548883 |
- |
- |
PREDICTED: probable nucleoredoxin 3 isoform X1 [Jatropha curcas] |