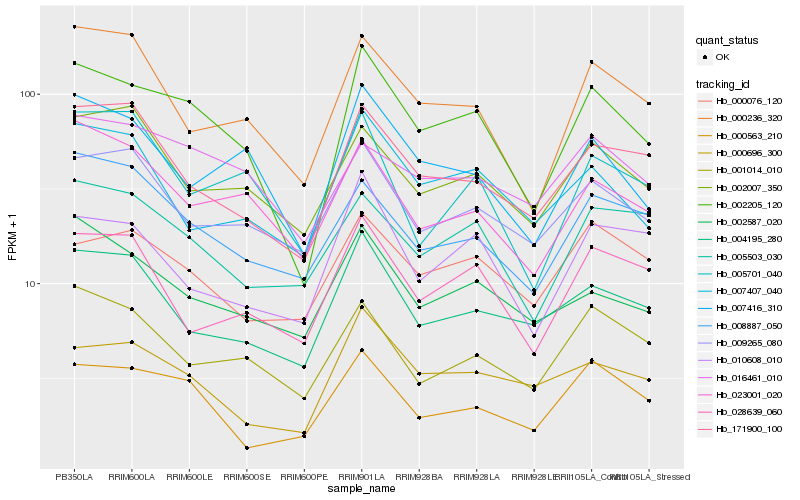
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_120 |
0.0 |
- |
- |
PREDICTED: O-acetyl-ADP-ribose deacetylase MACROD2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007416_310 |
0.1152193503 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 3 |
Hb_000236_320 |
0.1212792171 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 4 |
Hb_171900_100 |
0.124403347 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0270496 [Jatropha curcas] |
| 5 |
Hb_000696_300 |
0.1268578675 |
- |
- |
hypothetical protein EUGRSUZ_J01717 [Eucalyptus grandis] |
| 6 |
Hb_023001_020 |
0.1274647633 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 7 |
Hb_000563_210 |
0.1290776818 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 20a [Jatropha curcas] |
| 8 |
Hb_007407_040 |
0.13092341 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_002587_020 |
0.1326994376 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_004195_280 |
0.1329626032 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 11 |
Hb_009265_080 |
0.1336648866 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 12 |
Hb_001014_010 |
0.1349698588 |
- |
- |
PREDICTED: uncharacterized protein LOC105647311 [Jatropha curcas] |
| 13 |
Hb_008887_050 |
0.1360327617 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 14 |
Hb_002007_350 |
0.1360887343 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 15 |
Hb_010608_010 |
0.1363166138 |
- |
- |
PREDICTED: calcium-dependent protein kinase 3 [Jatropha curcas] |
| 16 |
Hb_005503_030 |
0.1367196088 |
- |
- |
PREDICTED: protein SCO1 homolog 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_016461_010 |
0.1375003019 |
- |
- |
PREDICTED: proline synthase co-transcribed bacterial homolog protein [Jatropha curcas] |
| 18 |
Hb_028639_060 |
0.1377577662 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_000076_120 |
0.1390335596 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 20 |
Hb_005701_040 |
0.1395396429 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |