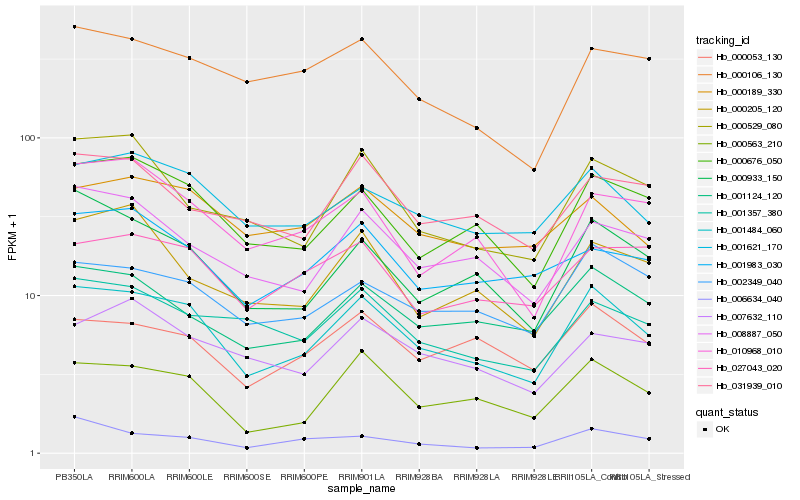
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001484_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641630 [Jatropha curcas] |
| 2 |
Hb_000563_210 |
0.0912759898 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 20a [Jatropha curcas] |
| 3 |
Hb_000676_050 |
0.0926135151 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B-like [Gossypium raimondii] |
| 4 |
Hb_001621_170 |
0.1004270387 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000933_150 |
0.1018433777 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001124_120 |
0.1047752648 |
- |
- |
PREDICTED: uncharacterized protein LOC105631831 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002349_040 |
0.1064015898 |
- |
- |
PREDICTED: RNA pseudouridine synthase 5 [Jatropha curcas] |
| 8 |
Hb_008887_050 |
0.1076656697 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 9 |
Hb_001357_380 |
0.1170913602 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_000053_130 |
0.1174868373 |
- |
- |
PREDICTED: uncharacterized protein At1g26090, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000529_080 |
0.117534736 |
- |
- |
PREDICTED: uncharacterized protein LOC105641675 [Jatropha curcas] |
| 12 |
Hb_000189_330 |
0.1183134824 |
- |
- |
PREDICTED: ER membrane protein complex subunit 2 [Jatropha curcas] |
| 13 |
Hb_010968_010 |
0.1194745632 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 14 |
Hb_000106_130 |
0.1203645939 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_027043_020 |
0.1221970588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001983_030 |
0.1226106275 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |
| 17 |
Hb_031939_010 |
0.122665389 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_007632_110 |
0.1246492383 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 19 |
Hb_000205_120 |
0.1251109112 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_006634_040 |
0.1251405795 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |