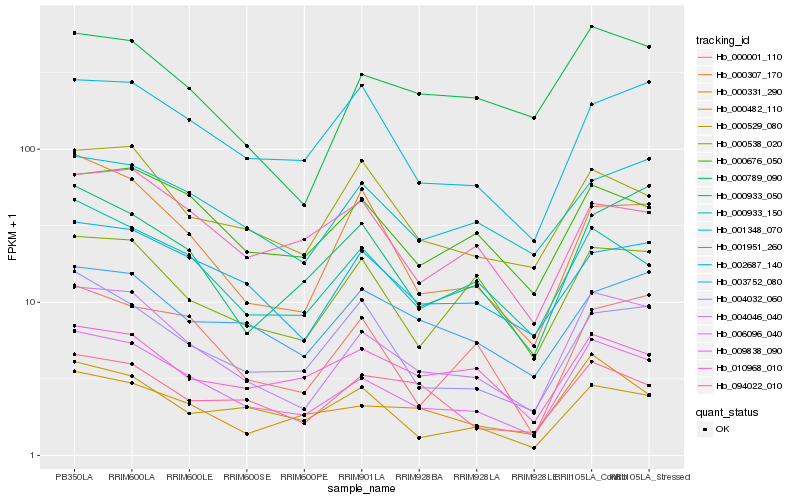
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006096_040 |
0.0 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 2 |
Hb_004046_040 |
0.0572107315 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 3 |
Hb_000933_150 |
0.1006231459 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000676_050 |
0.1164604186 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B-like [Gossypium raimondii] |
| 5 |
Hb_002687_140 |
0.1204897708 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 6 |
Hb_000482_110 |
0.1223315804 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000331_290 |
0.1235369999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004032_060 |
0.1245618605 |
- |
- |
elongation factor 1 gamma-like protein, partial [Ipomoea nil] |
| 9 |
Hb_000307_170 |
0.1297184263 |
- |
- |
PREDICTED: uncharacterized protein LOC105638528 [Jatropha curcas] |
| 10 |
Hb_010968_010 |
0.1300308541 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 11 |
Hb_000529_080 |
0.1306435387 |
- |
- |
PREDICTED: uncharacterized protein LOC105641675 [Jatropha curcas] |
| 12 |
Hb_000538_020 |
0.1334976965 |
- |
- |
PREDICTED: autophagy protein 5 [Jatropha curcas] |
| 13 |
Hb_001951_260 |
0.1335578933 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 14 |
Hb_001348_070 |
0.1338635257 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000933_050 |
0.136526592 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000001_110 |
0.1365792394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009838_090 |
0.1373246845 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000789_090 |
0.137413096 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 19 |
Hb_003752_080 |
0.1378717513 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 20 |
Hb_094022_010 |
0.1379246534 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |