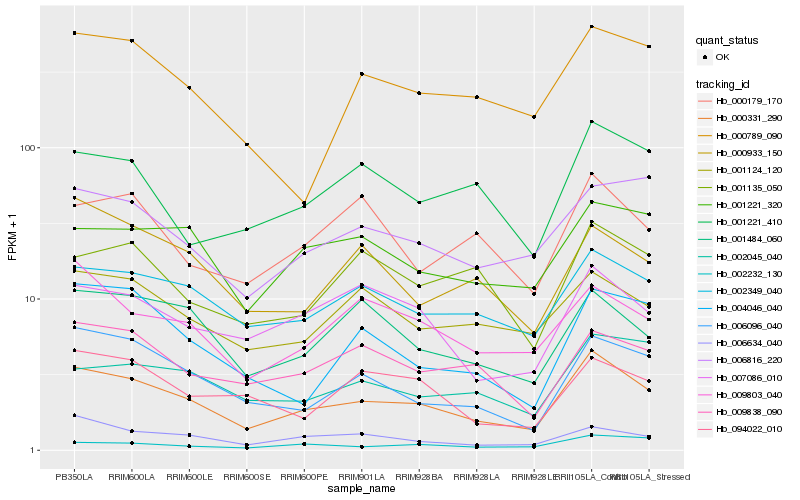
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_290 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002349_040 |
0.1162171875 |
- |
- |
PREDICTED: RNA pseudouridine synthase 5 [Jatropha curcas] |
| 3 |
Hb_006096_040 |
0.1235369999 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 4 |
Hb_006634_040 |
0.1353915961 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 5 |
Hb_001484_060 |
0.1367314811 |
- |
- |
PREDICTED: uncharacterized protein LOC105641630 [Jatropha curcas] |
| 6 |
Hb_001124_120 |
0.1393682345 |
- |
- |
PREDICTED: uncharacterized protein LOC105631831 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007086_010 |
0.1401816812 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26-like [Jatropha curcas] |
| 8 |
Hb_001221_320 |
0.1402957634 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_009838_090 |
0.140566918 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 10 |
Hb_006816_220 |
0.1425976337 |
- |
- |
PREDICTED: zinc finger matrin-type protein 2 [Jatropha curcas] |
| 11 |
Hb_009803_040 |
0.1433175057 |
- |
- |
tetratricopeptide repeat-containing family protein [Populus trichocarpa] |
| 12 |
Hb_004046_040 |
0.1456135077 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 13 |
Hb_002045_040 |
0.1459085253 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000789_090 |
0.1467457541 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 15 |
Hb_001221_410 |
0.1477561929 |
- |
- |
PREDICTED: gamma carbonic anhydrase-like 2, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000933_150 |
0.149367566 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_094022_010 |
0.1521473316 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |
| 18 |
Hb_002232_130 |
0.1526304203 |
- |
- |
- |
| 19 |
Hb_001135_050 |
0.1527741627 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000179_170 |
0.1548765868 |
- |
- |
PREDICTED: protein bem46 isoform X1 [Jatropha curcas] |